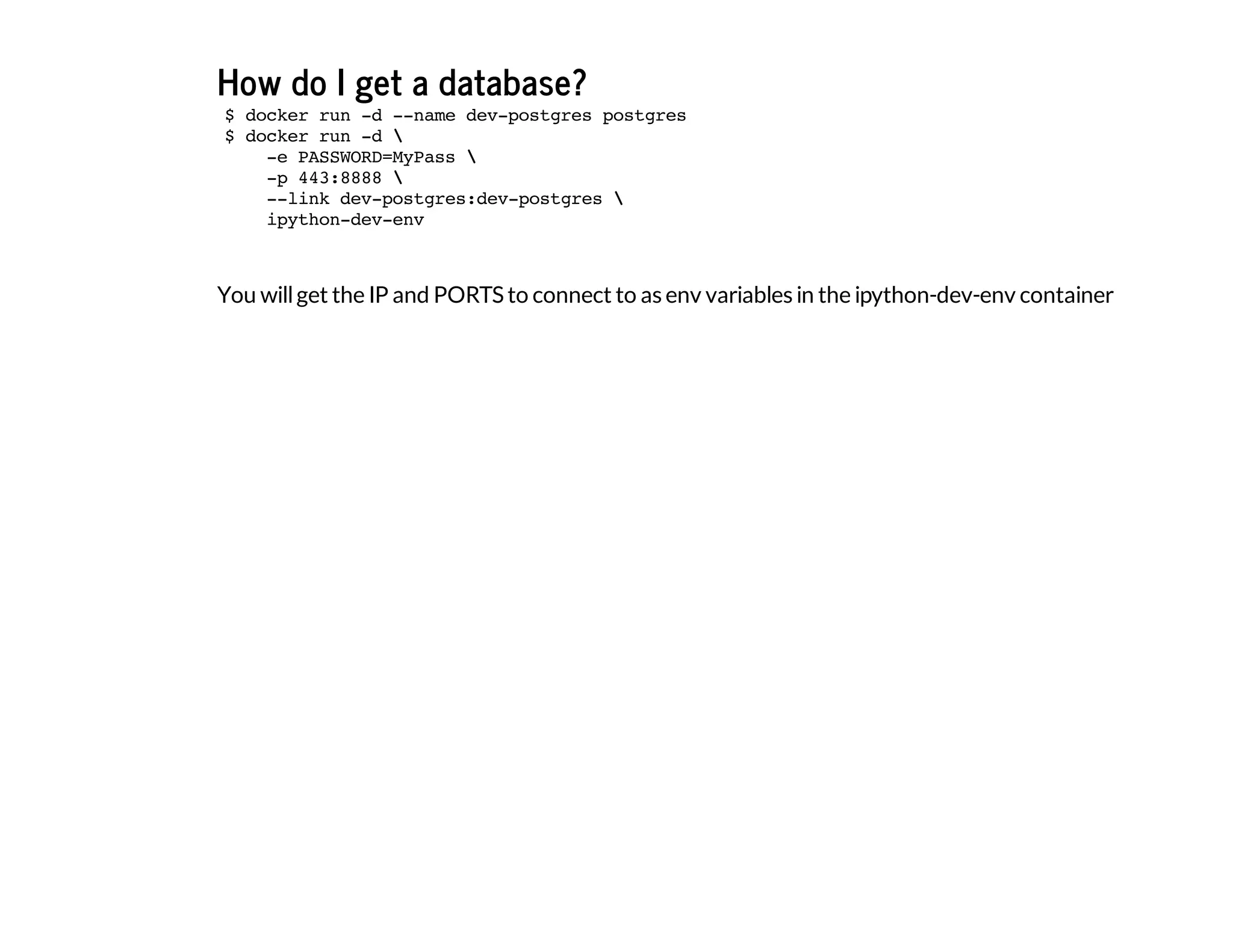
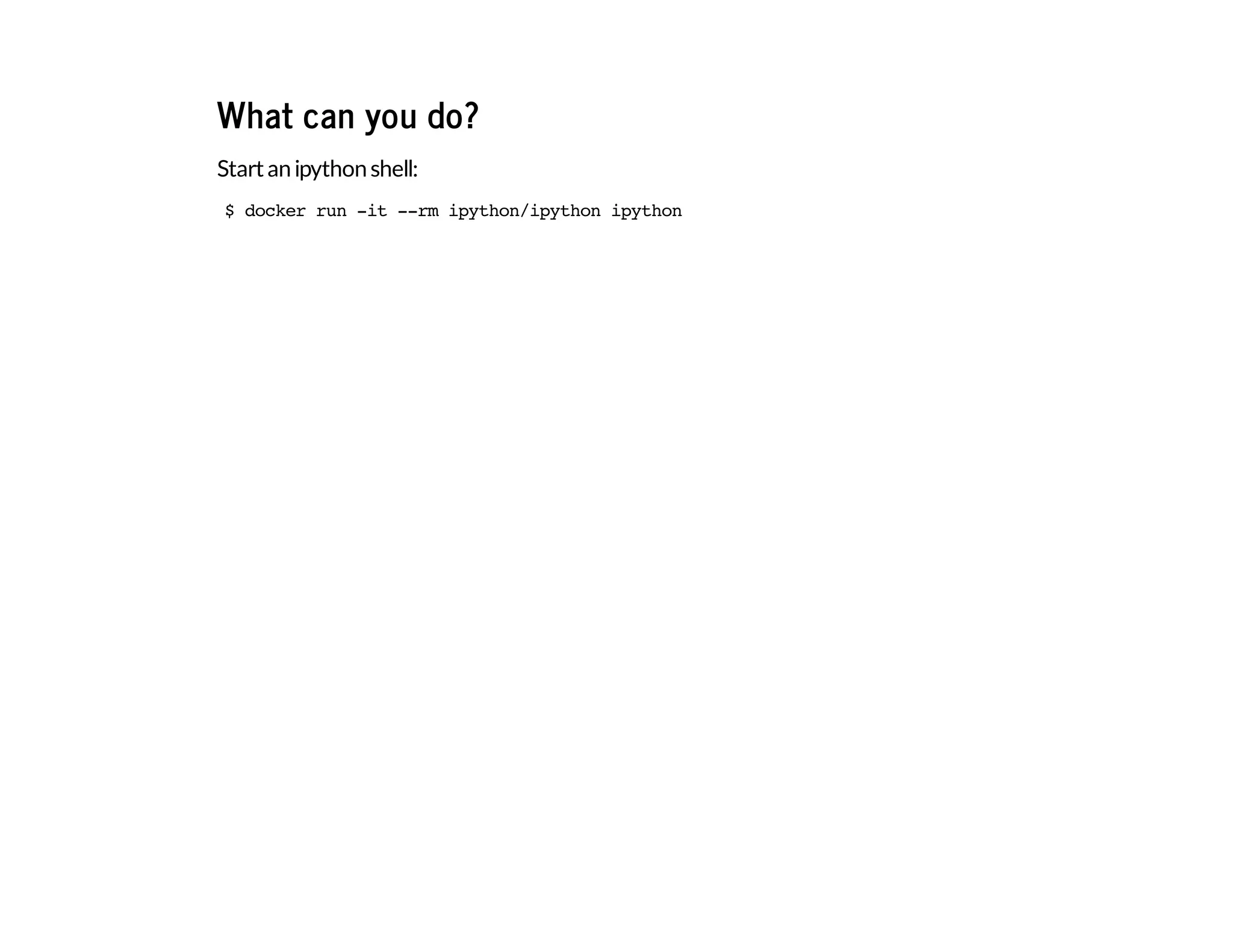
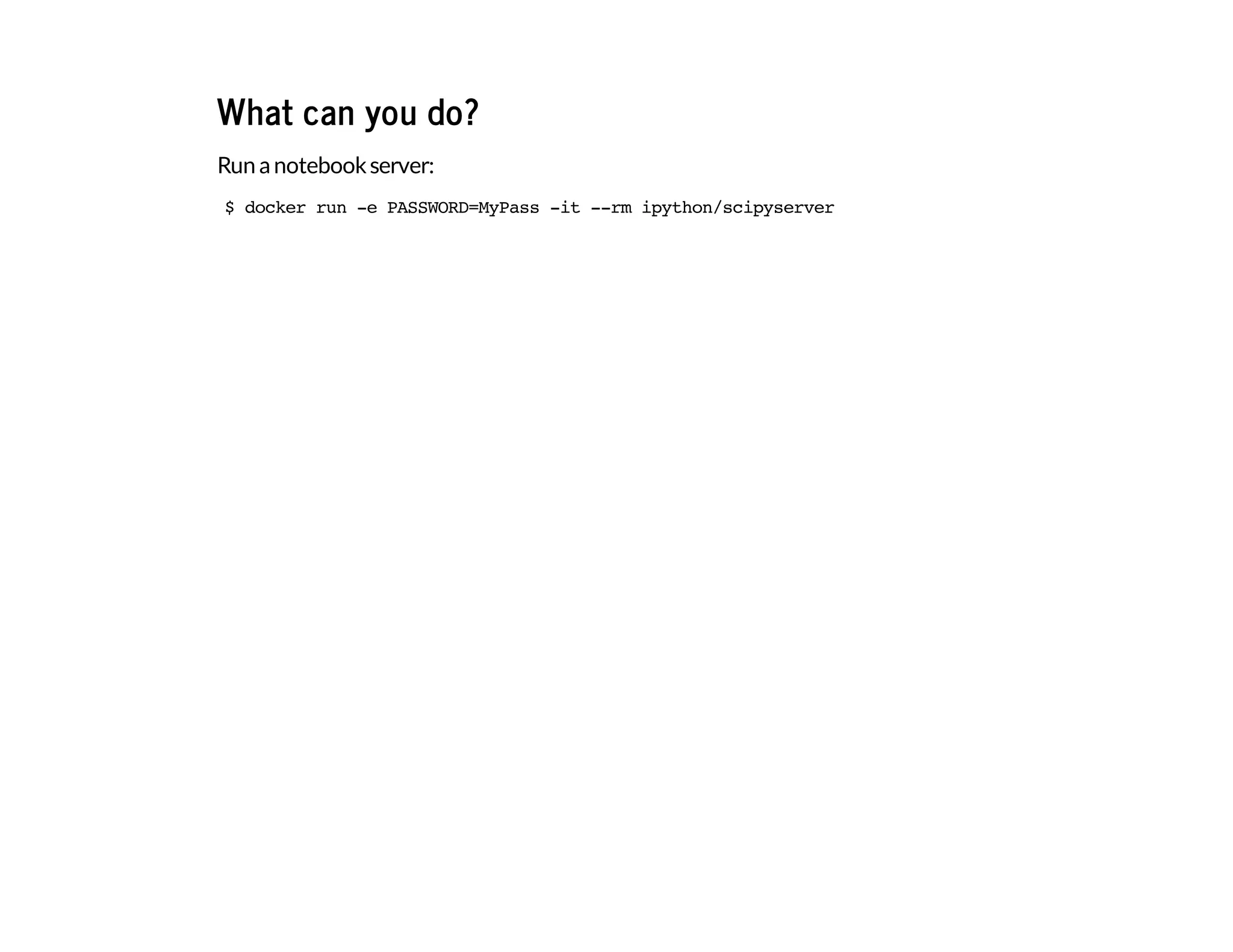
The document discusses the use of Docker for data science, highlighting its advantages over traditional package managers and setups. It details a personal journey of transitioning from a complicated development setup to a streamlined Docker environment, showcasing commands and configurations for running Python data science tools within Docker containers. The author emphasizes Docker's portability, ease of configuration, and ability to manage multiple independent environments efficiently.


























![Install an extra python module into a notebook server
Test the installof the package you want:
In[5]: !pip3searchgensim
gensim -PythonframeworkforfastVectorSpaceModelling
In[7]: !pip3installgensim
Downloading/unpackinggensim
Downloadinggensim-0.10.3.tar.gz(3.1MB):3.1MBdownloaded
Runningsetup.py(path:/tmp/pip_build_root/gensim/setup.py)egg_infoforpackagegensim
warning:nofilesfoundmatching'*.sh'underdirectory'.'
nopreviously-includeddirectoriesfoundmatching'docs/src*'
Requirementalreadysatisfied(use--upgradetoupgrade):numpy>=1.3in/usr/local/lib/pyt
hon2.7/dist-packages(fromgensim)
Requirementalreadysatisfied(use--upgradetoupgrade):scipy>=0.7.0in/usr/local/lib/p
ython2.7/dist-packages(fromgensim)
Requirementalreadysatisfied(use--upgradetoupgrade):six>=1.2.0in/usr/lib/python2.7
/dist-packages(fromgensim)
Installingcollectedpackages:gensim
Runningsetup.pyinstallforgensim
warning:nofilesfoundmatching'*.sh'underdirectory'.'
nopreviously-includeddirectoriesfoundmatching'docs/src*'
building'gensim.models.word2vec_inner'extension
x86_64-linux-gnu-gcc-pthread-fno-strict-aliasing-DNDEBUG-g-fwrapv-O2-Wall-Wstr
ict-prototypes-fPIC-I/tmp/pip_build_root/gensim/gensim/models-I/usr/include/python2.7-
I/usr/local/lib/python2.7/dist-packages/numpy/core/include-c./gensim/models/word2vec_inn
er.c-obuild/temp.linux-x86_64-2.7/./gensim/models/word2vec_inner.o
Infileincludedfrom/usr/include/python2.7/numpy/ndarraytypes.h:1761:0,
from/usr/include/python2.7/numpy/ndarrayobject.h:17,
from/usr/include/python2.7/numpy/arrayobject.h:4,
from./gensim/models/word2vec_inner.c:232:
/usr/include/python2.7/numpy/npy_1_7_deprecated_api.h:15:2:warning:#warning"Usingd
eprecatedNumPyAPI,disableitby""#definingNPY_NO_DEPRECATED_APINPY_1_7_API_VERSION"
[-Wcpp]](https://image.slidesharecdn.com/dockerfordatascienceslides-150108045743-conversion-gate01/75/Docker-for-data-science-30-2048.jpg)
![In [10]: import gensim
Ifthatworks,addtheinstallcommandstoyourDockerfile:
Andrebuild:
FROMipython/scipyserver
RUNpip2installgensim
RUNpip3installgensim
$dockerbuild-tipython-dev-env.
$dockerrun-i-t--rm-ePASSWORD=MyPass-p443:8888ipython-dev-env](https://image.slidesharecdn.com/dockerfordatascienceslides-150108045743-conversion-gate01/75/Docker-for-data-science-31-2048.jpg)

![But what do I put in requirements.txt?
In[12]: !pip3freeze|head
Cython==0.20.1post0
Jinja2==2.7.2
MarkupSafe==0.18
Pillow==2.3.0
Pygments==1.6
SQLAlchemy==0.9.8
Sphinx==1.2.2
brewer2mpl==1.4.1
certifi==14.05.14
chardet==2.0.1](https://image.slidesharecdn.com/dockerfordatascienceslides-150108045743-conversion-gate01/75/Docker-for-data-science-33-2048.jpg)