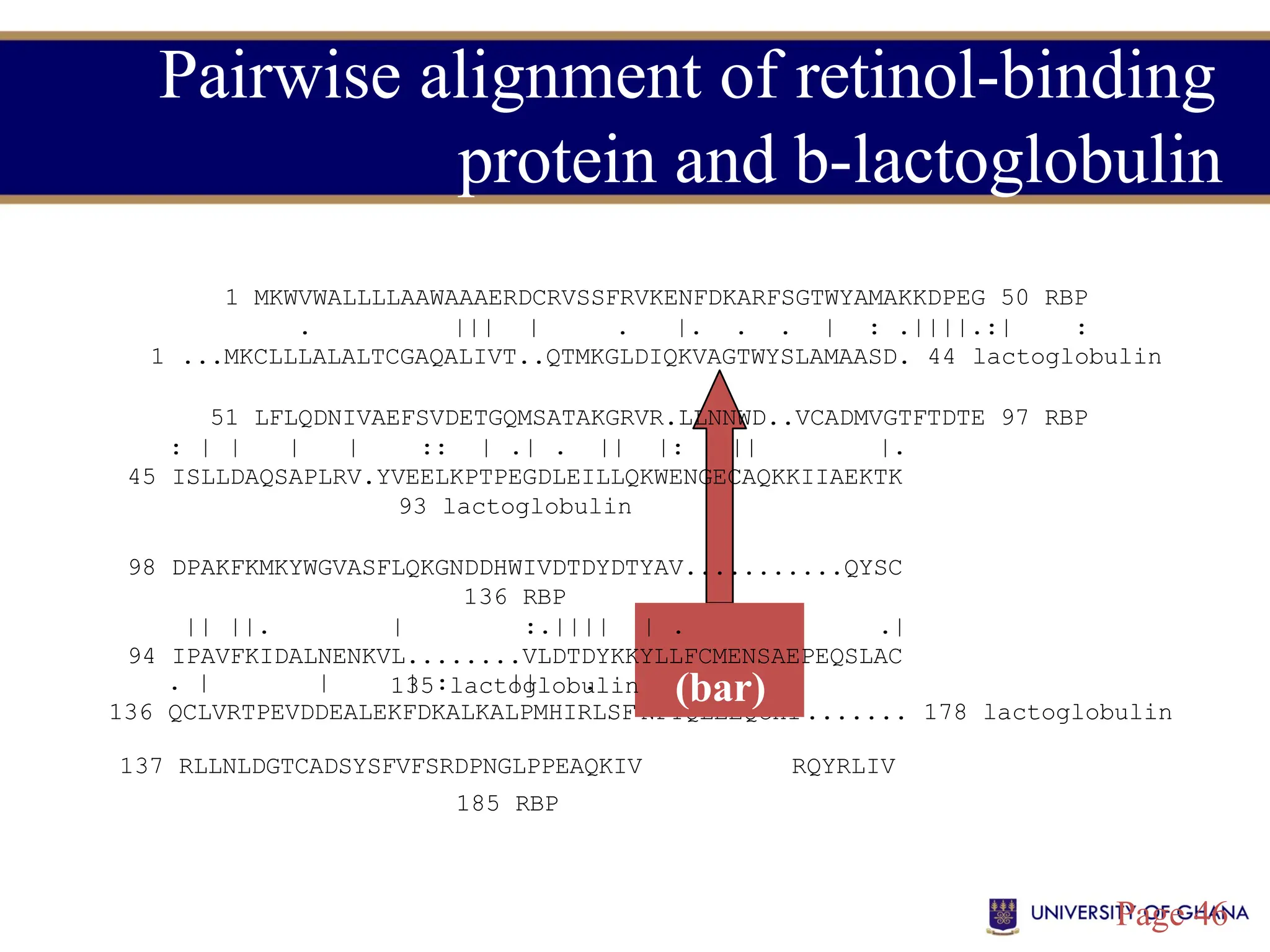
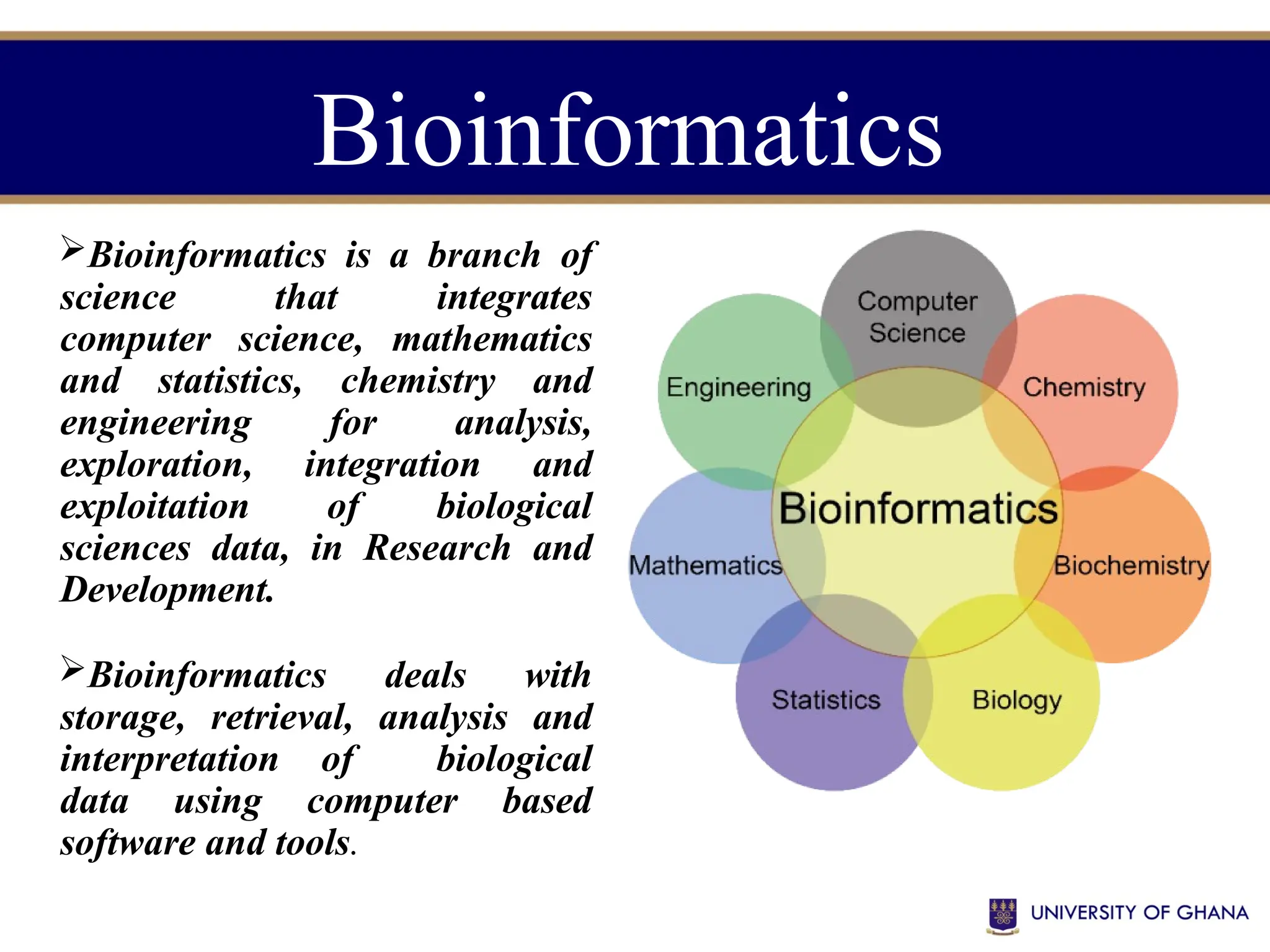
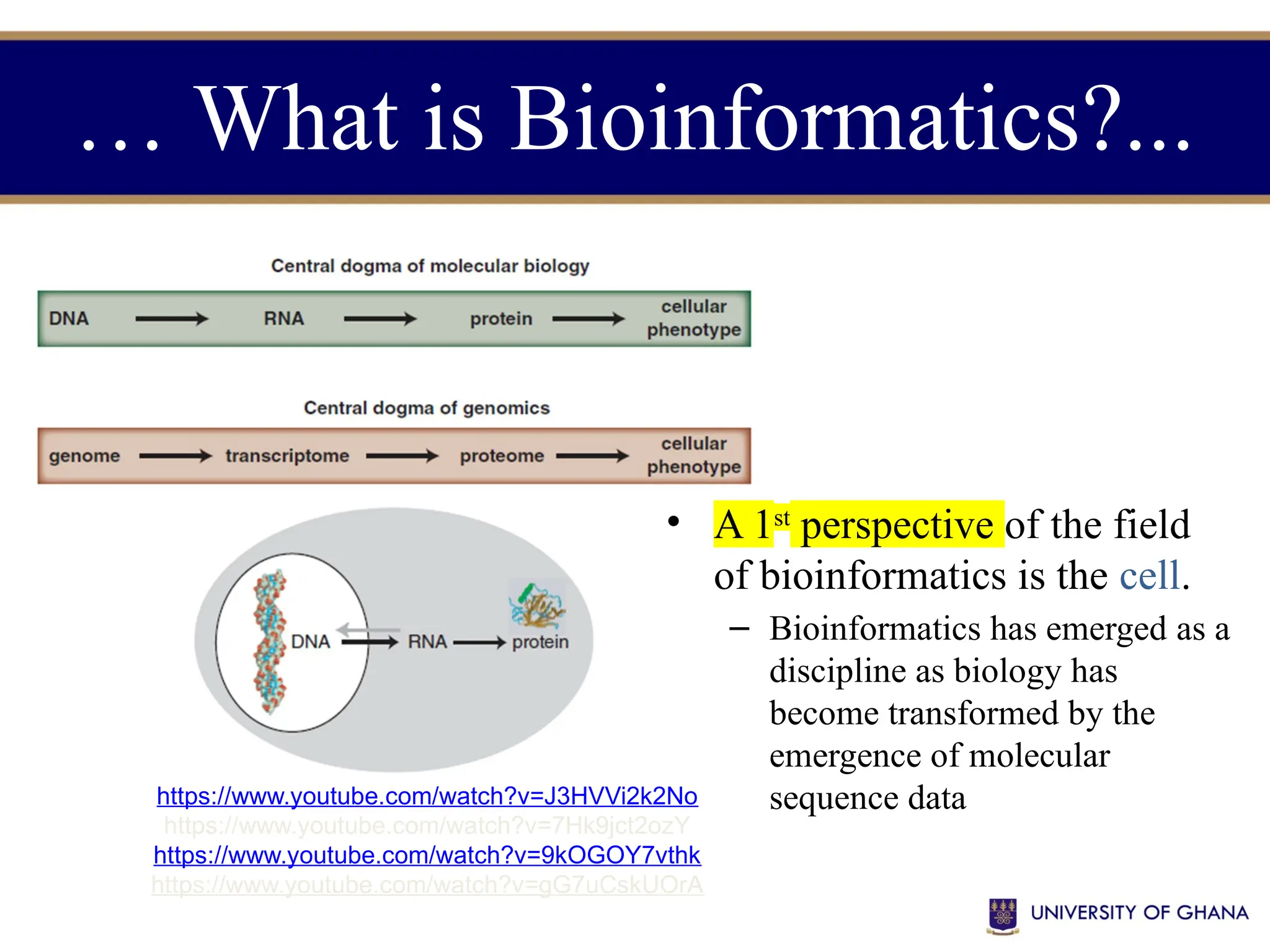
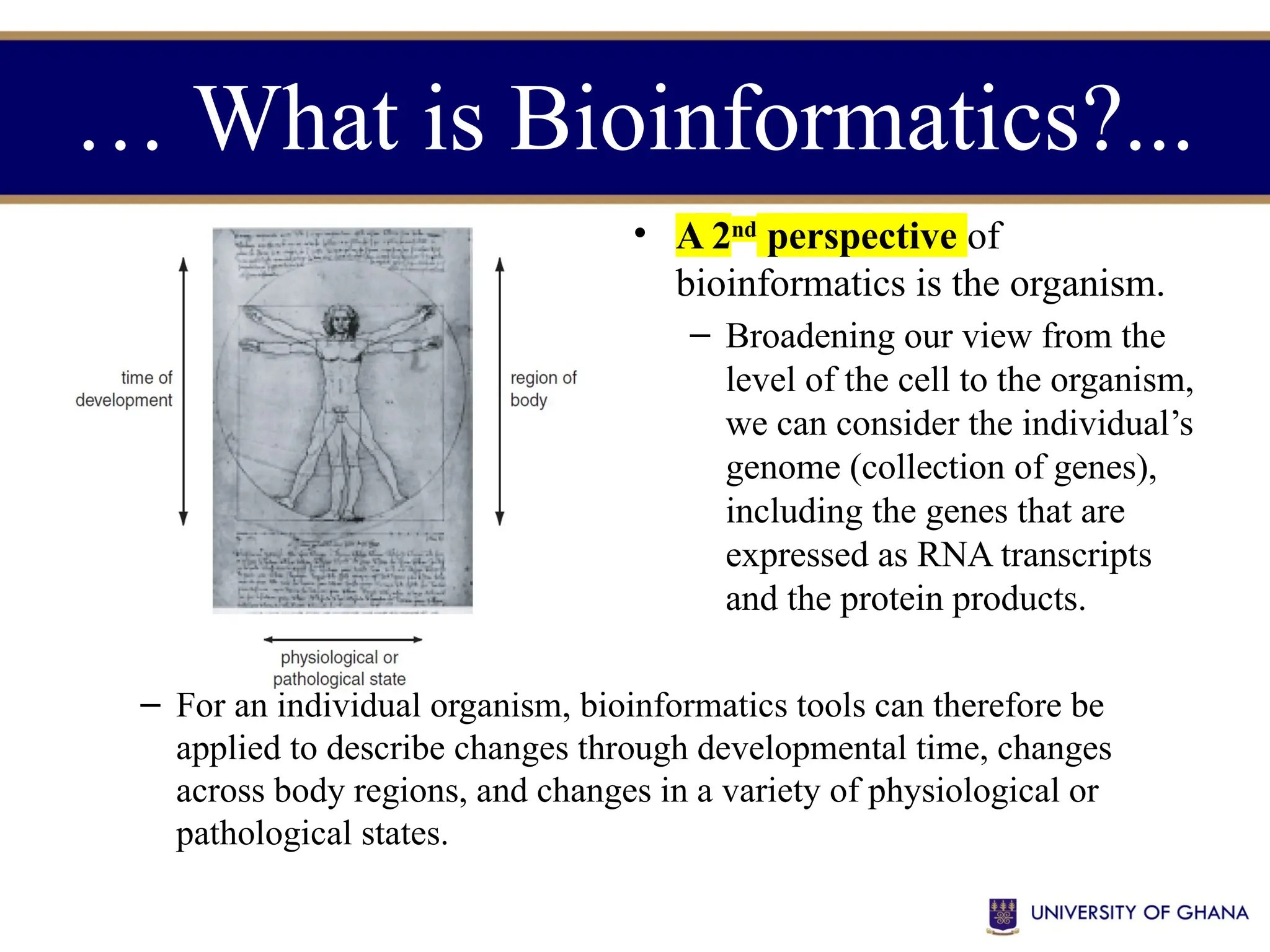
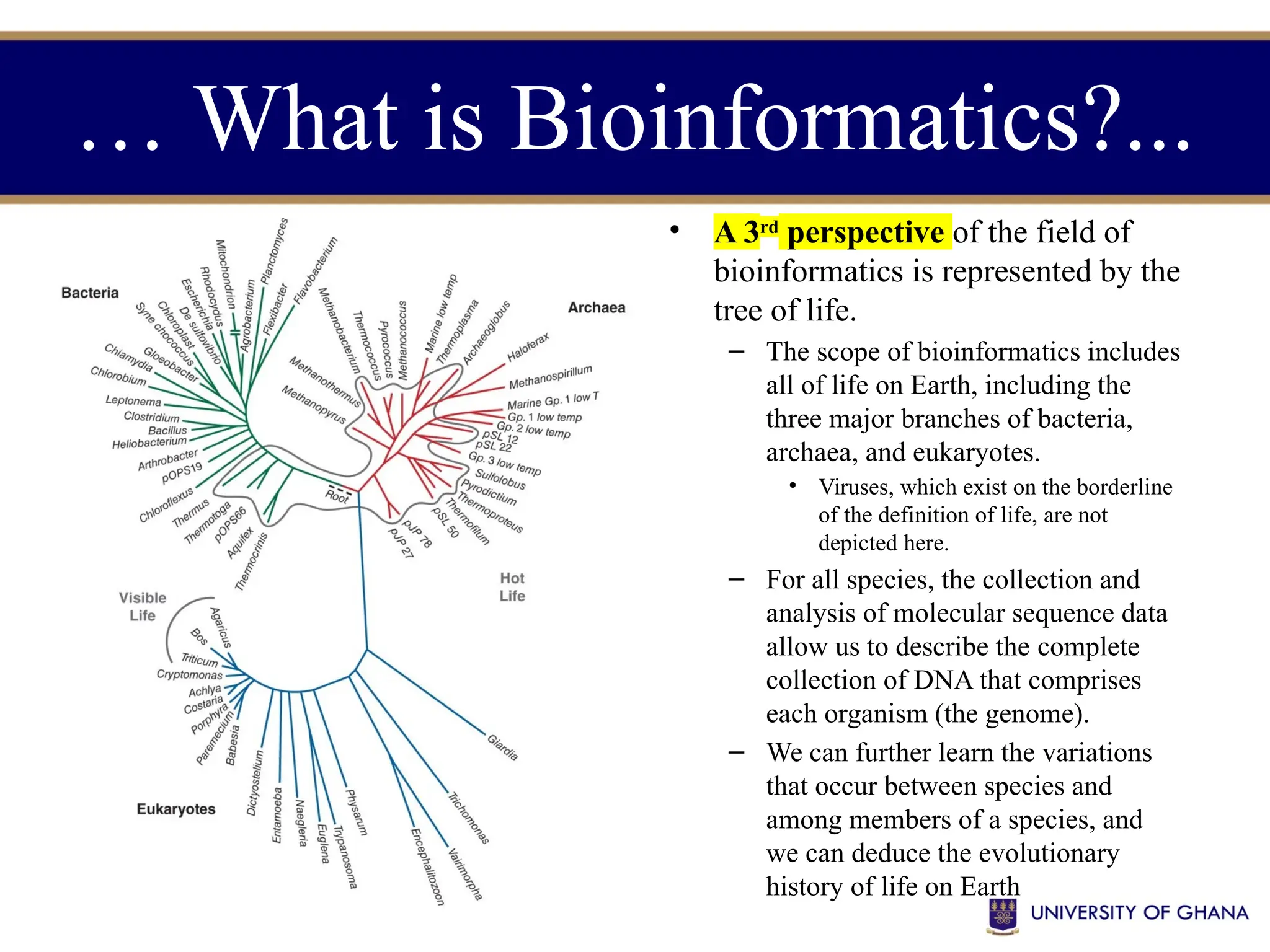
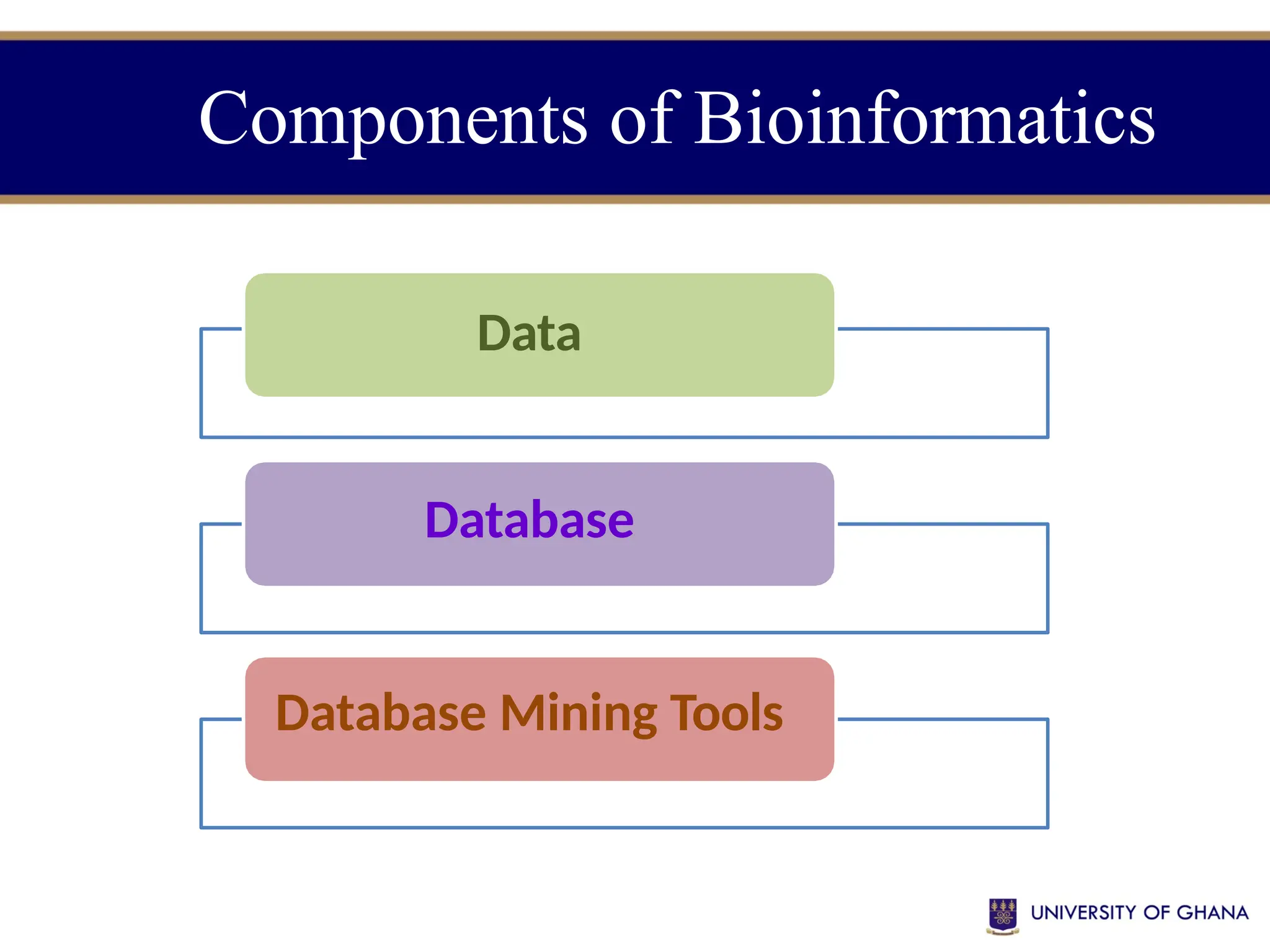
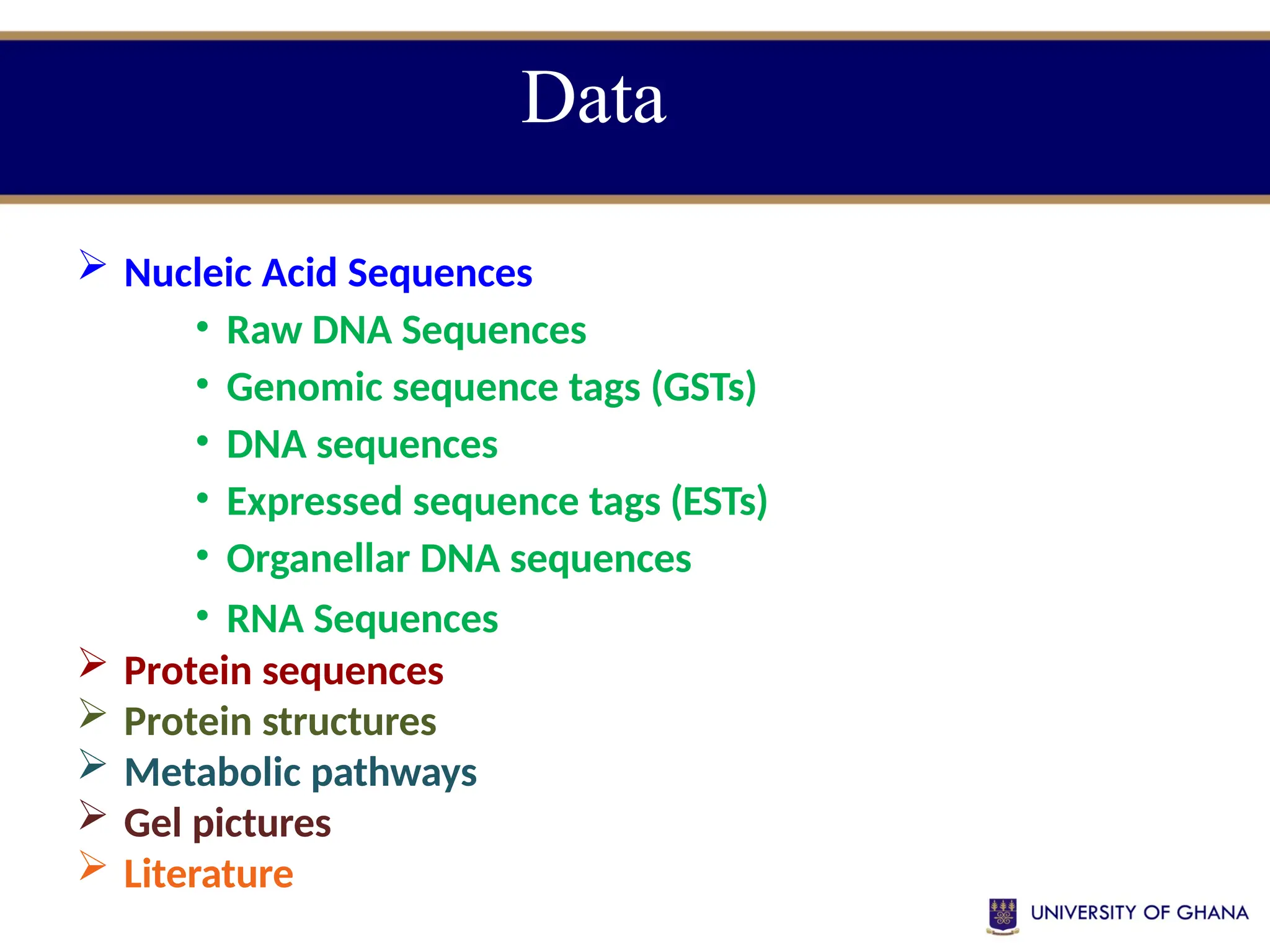
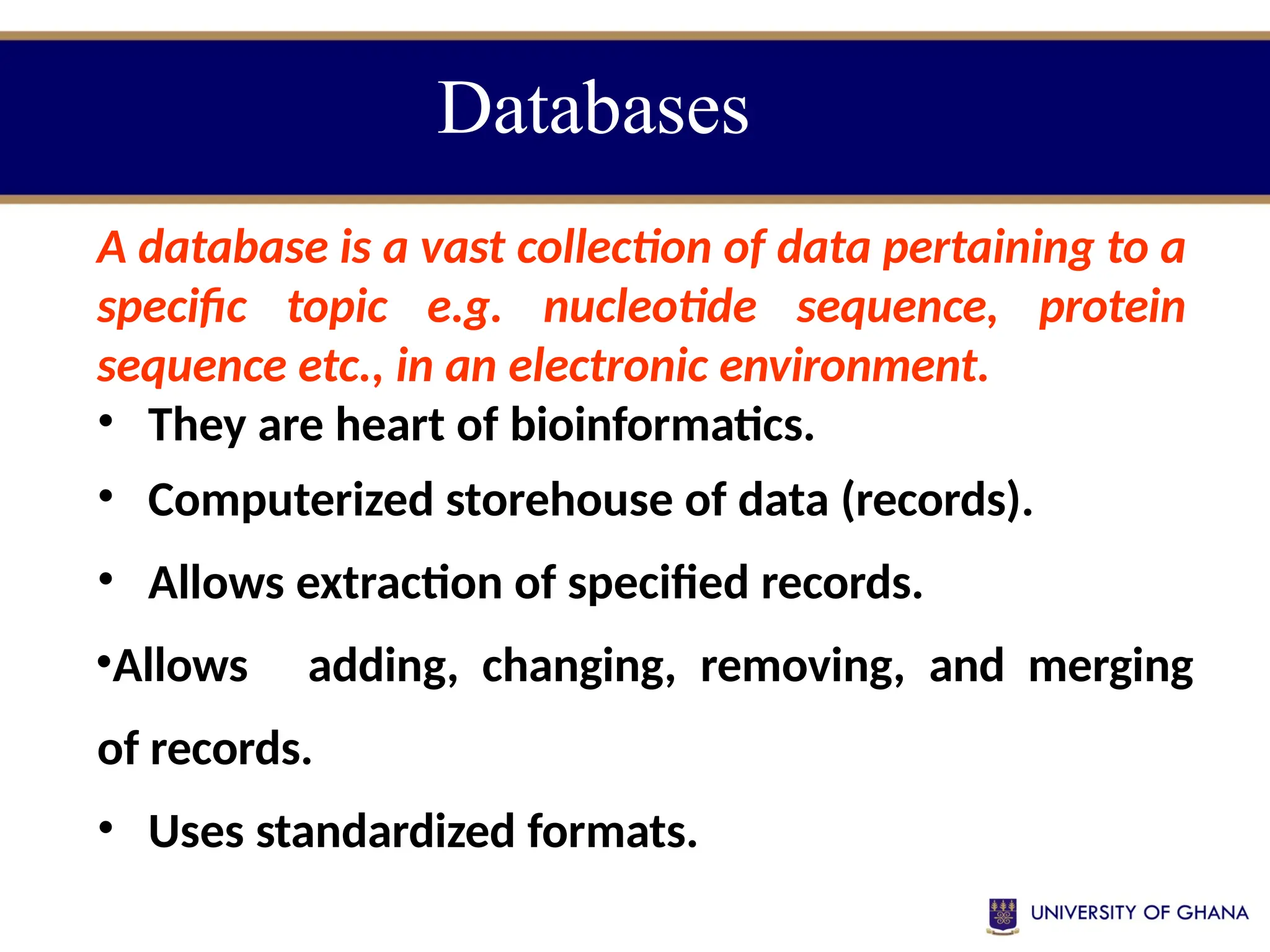
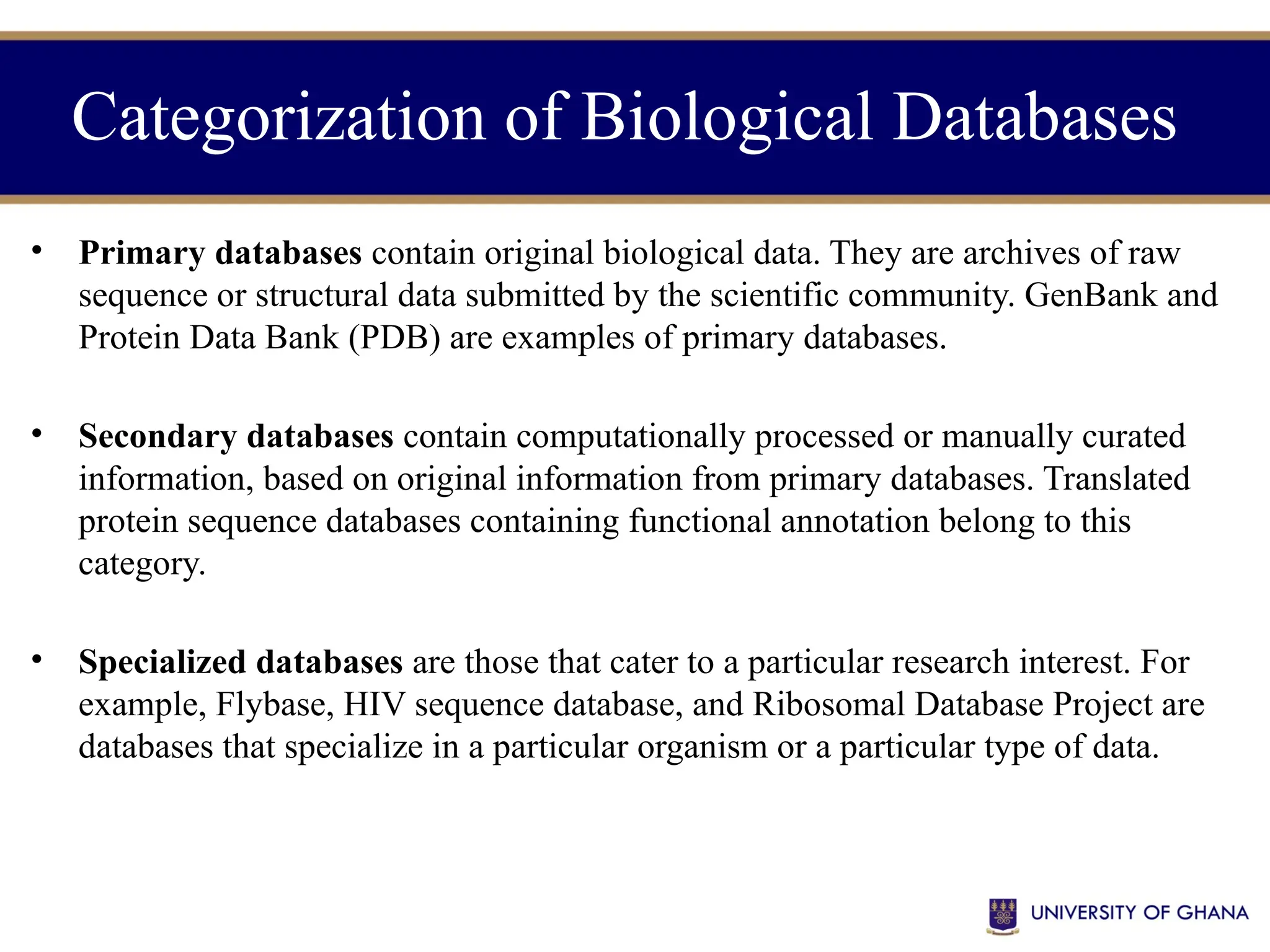
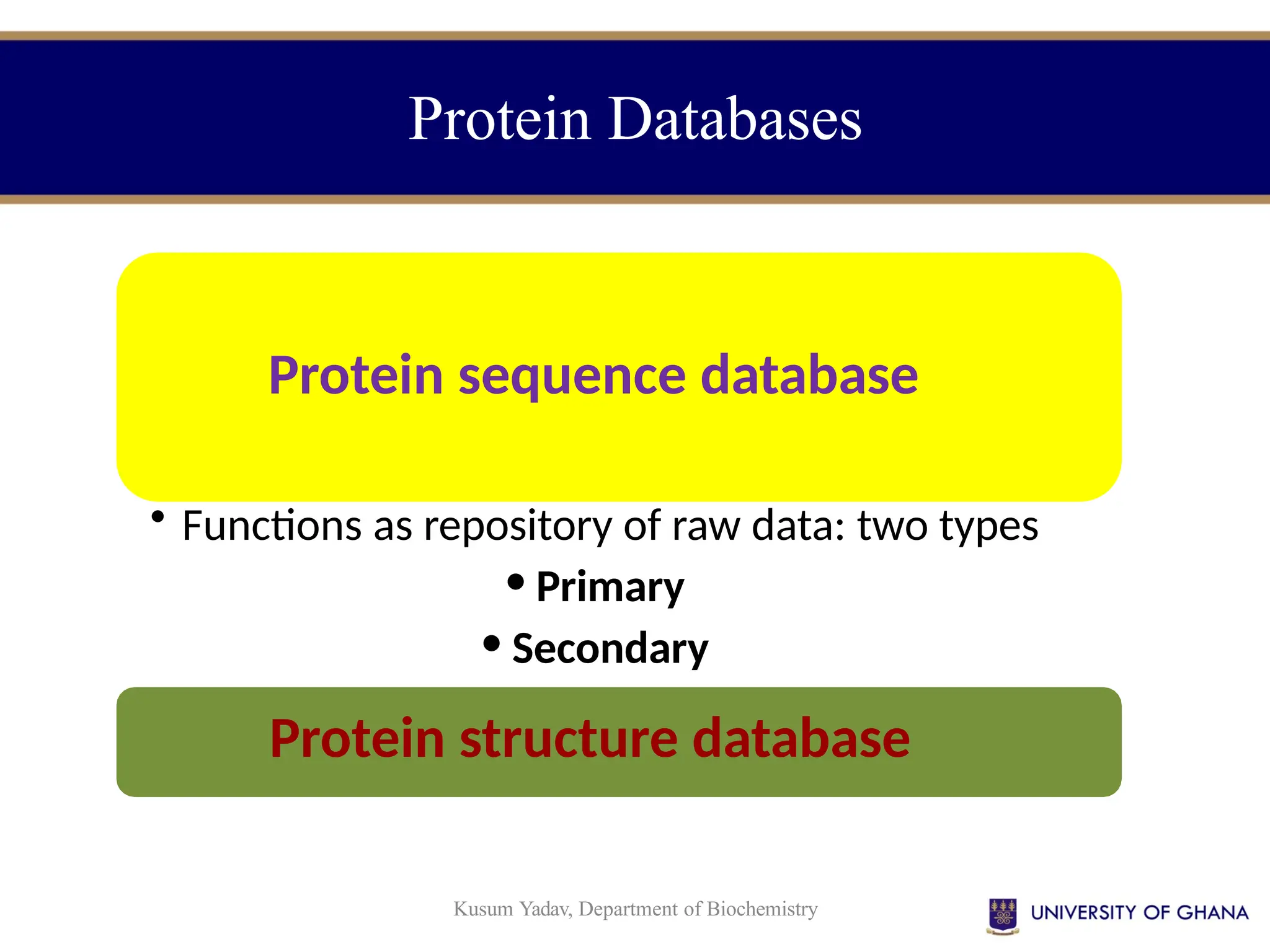
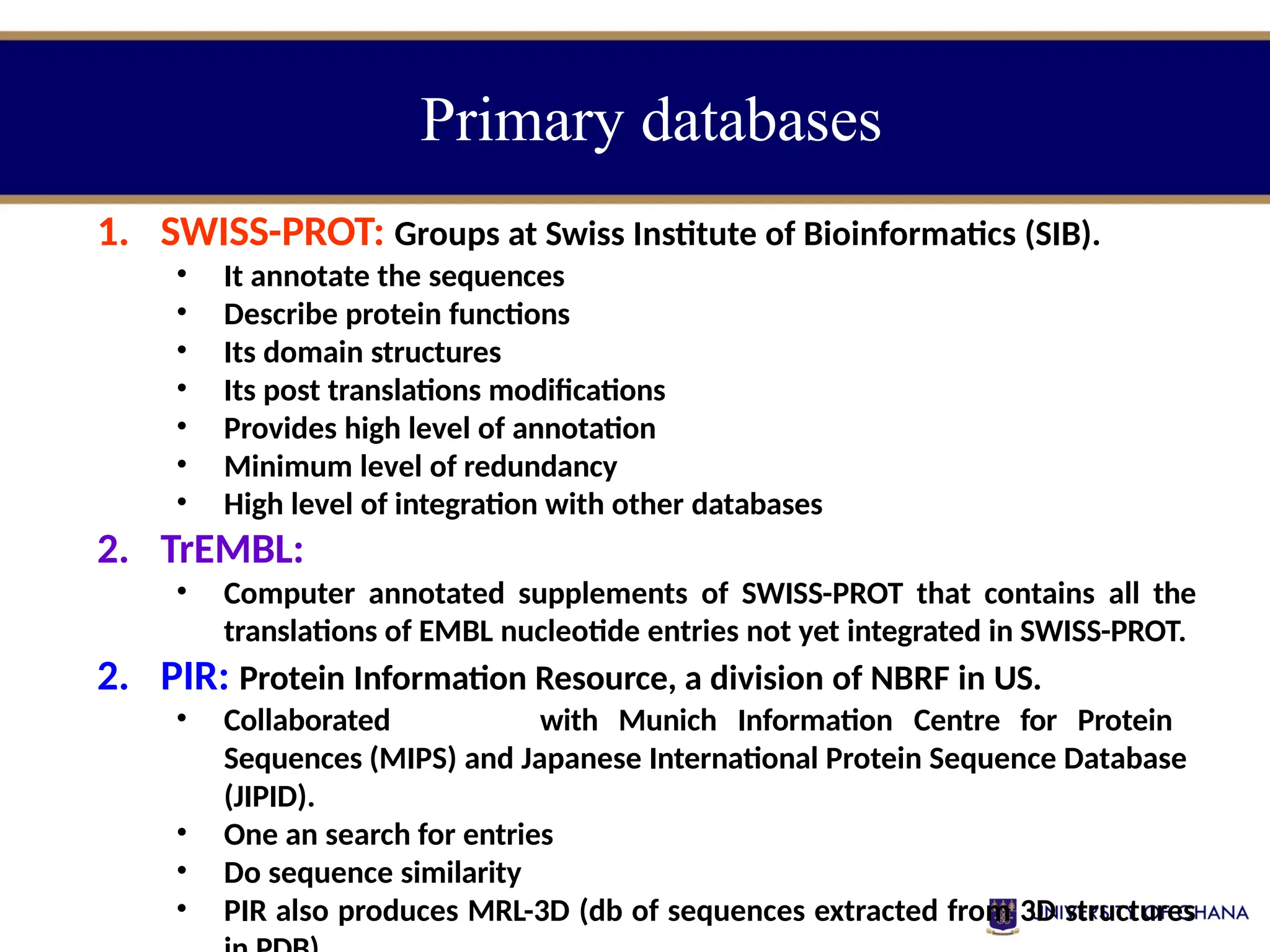
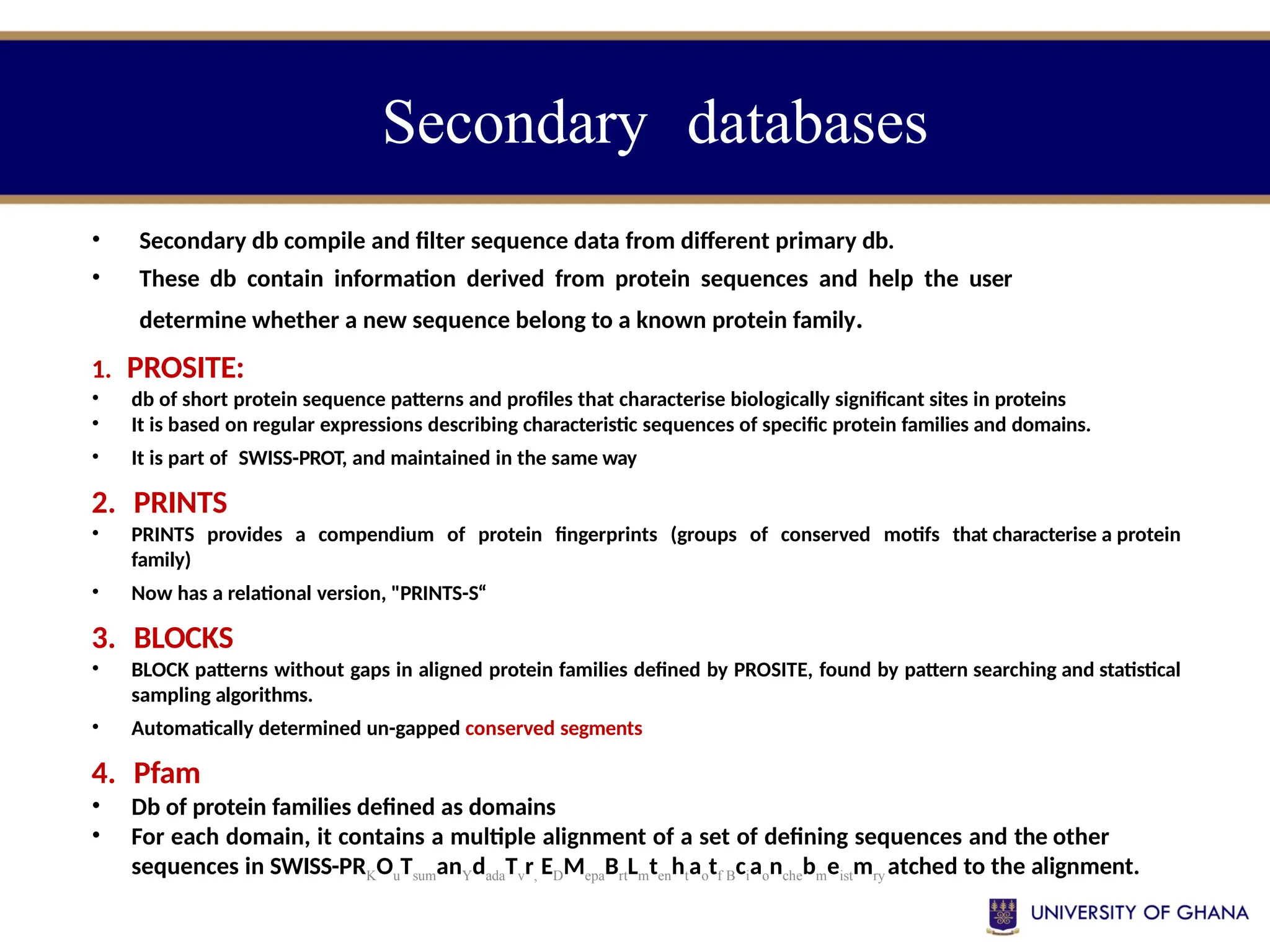
The document provides an introduction to bioinformatics, outlining its integration of computer science, mathematics, and biological data analysis to enhance understanding of biological processes. It covers the historical development of the field, various perspectives of bioinformatics, its key goals and applications, as well as necessary skills and tools. The document also highlights the importance of databases and the limitations of bioinformatics predictions, emphasizing the need for rigorous computational methods and a comprehensive understanding of both biology and data science.

![…What is Bioinformatics?...
• has been used for in silico analyses of biological queries
using mathematical and statistical techniques.
• [In silico (Latin for "in silicon") is an expression
used to mean "performed on computer or via
computer simulation.]
• primary goal is to increase the understanding of biological
processes.
• focuses on developing and applying computationally
intensive techniques to achieve this goal.](https://image.slidesharecdn.com/lecture1introductionbioinformatics-241203112510-fbe24d4e/75/Lecture_1_Introduction_Bioinformatics-pptx-9-2048.jpg)

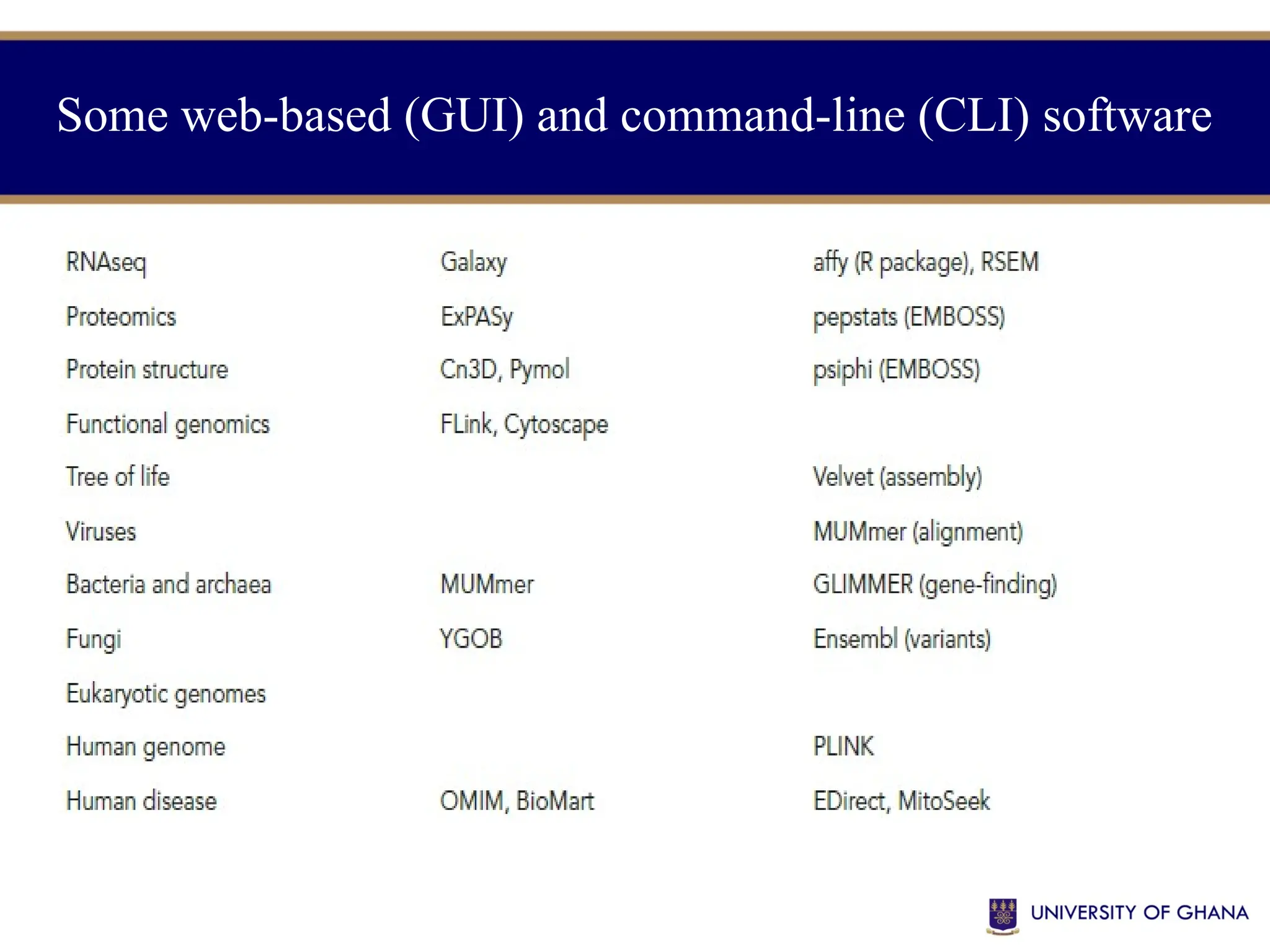



![Enzyme Database
BRENDA [BRaunshchweig ENzyme DAtabase]
Enzyme, a part of ExPaSy (Expert
Protein Analysis System, the proteomic
server of Swiss Institute of Bioinformatics)](https://image.slidesharecdn.com/lecture1introductionbioinformatics-241203112510-fbe24d4e/75/Lecture_1_Introduction_Bioinformatics-pptx-53-2048.jpg)