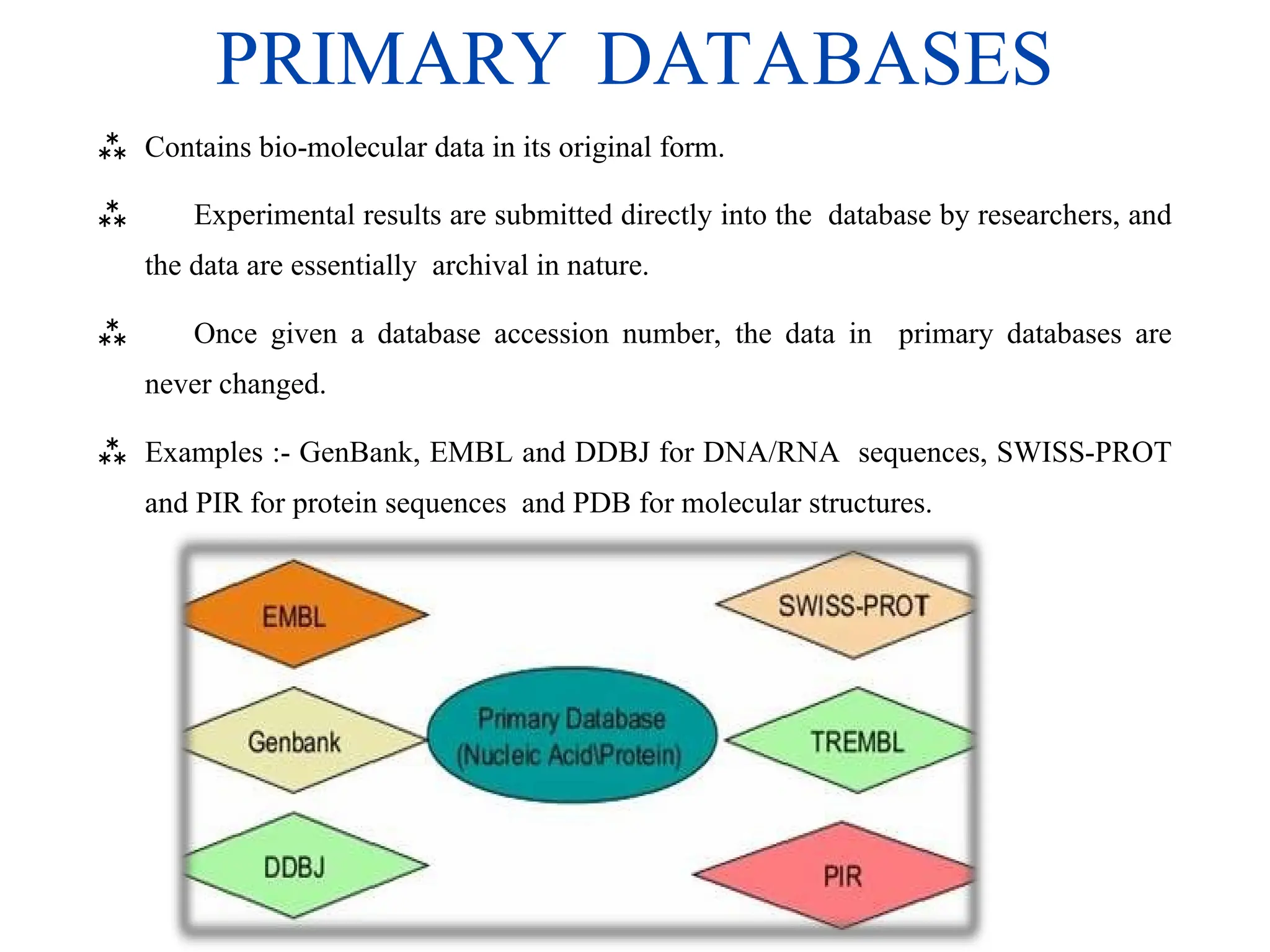
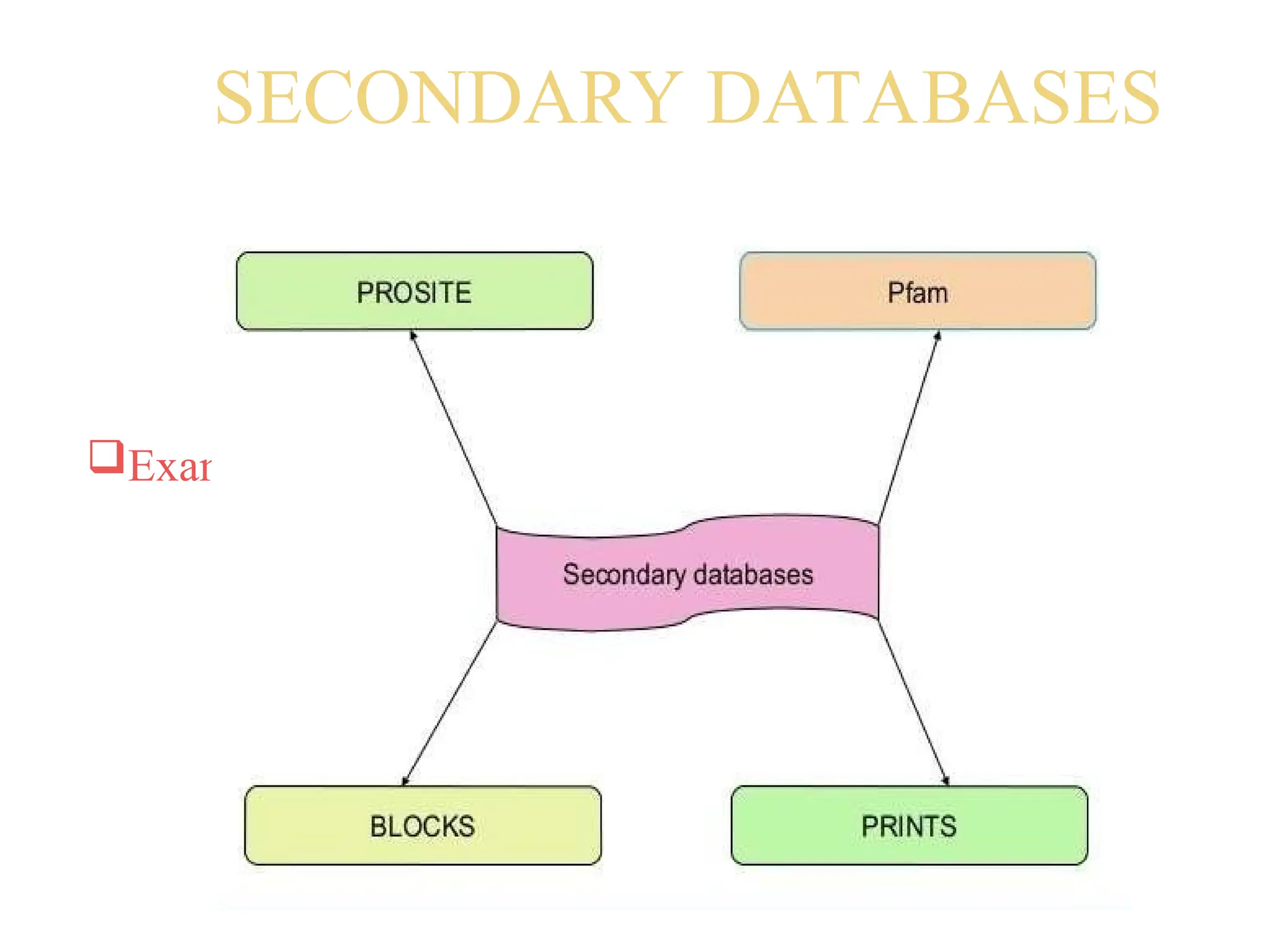
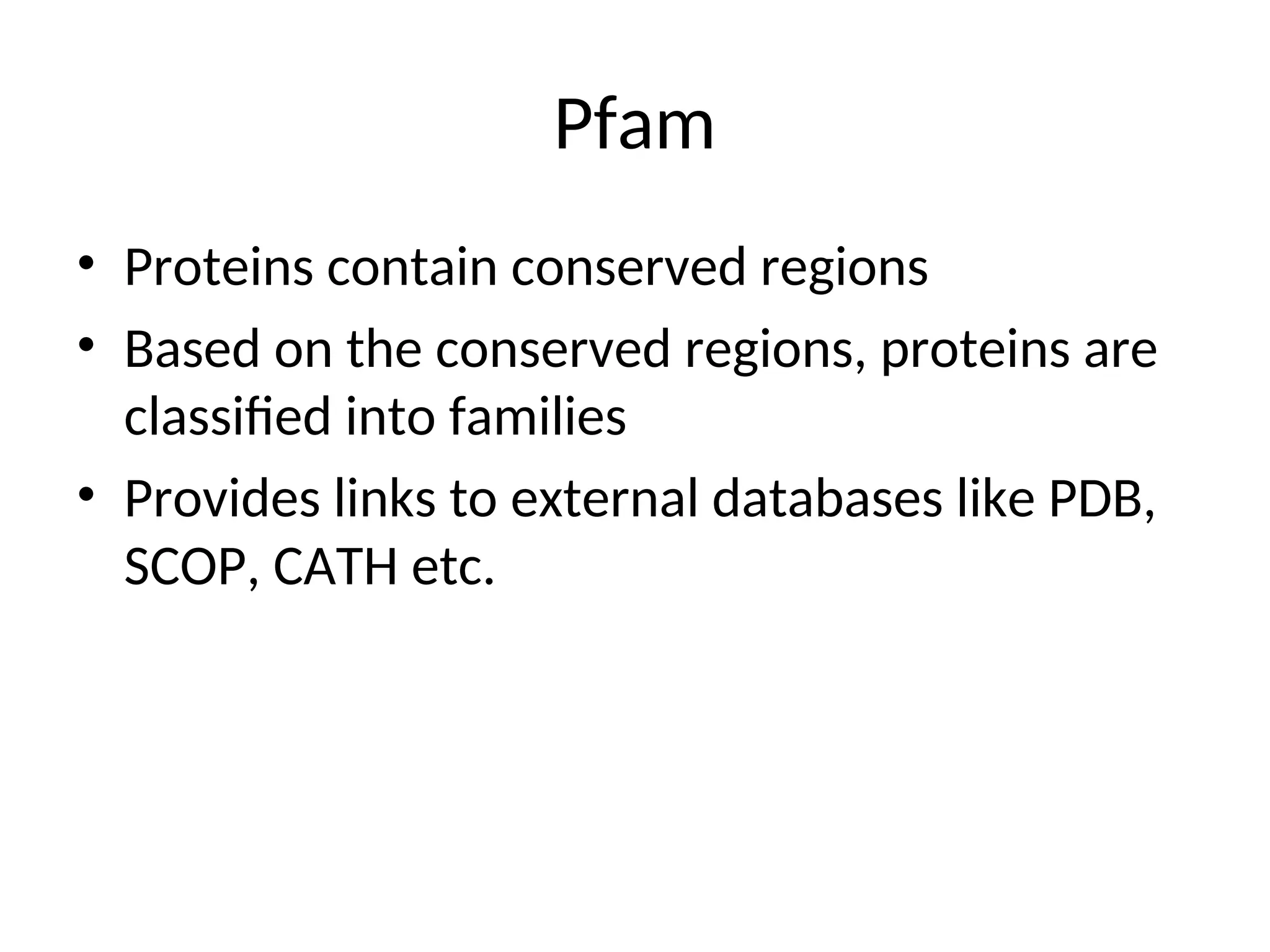
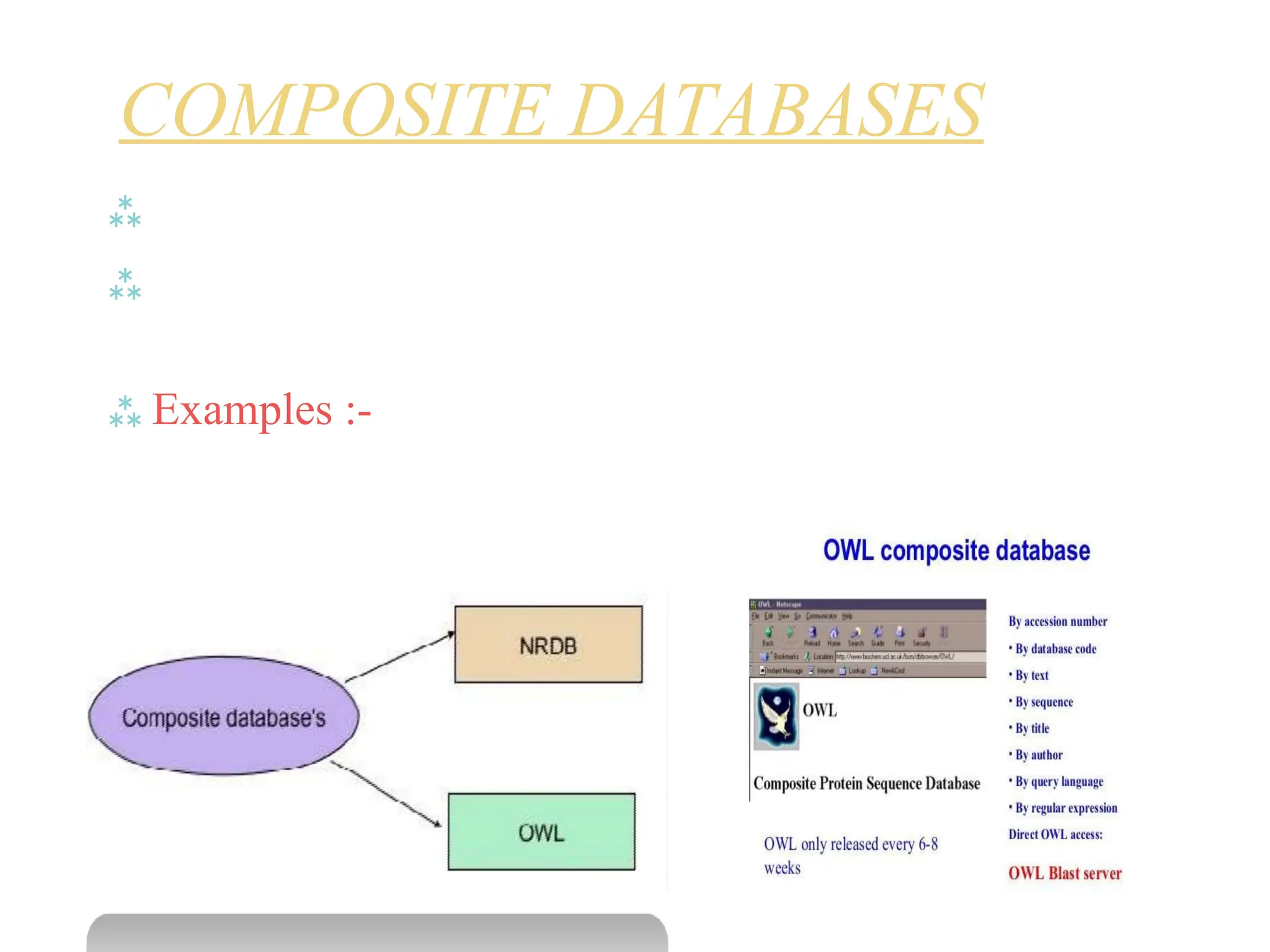
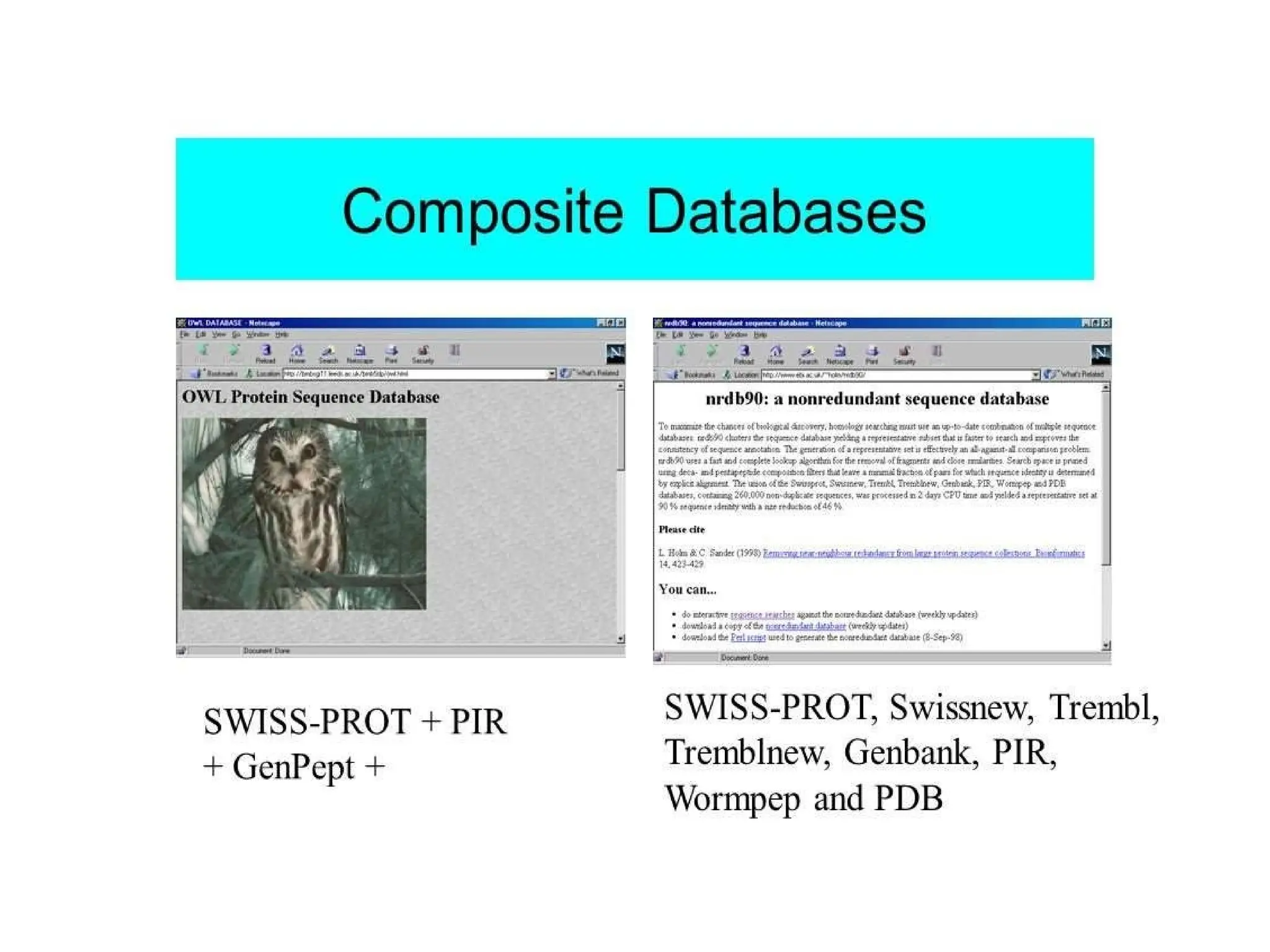
The document provides a comprehensive overview of various biological databases in molecular biology, detailing primary, secondary, and composite databases like GenBank, UniProt, and PDB. It outlines expectations from databases, their structures, and types of data they contain, emphasizing the importance of well-maintained and cross-referenced databases for biological research. Additionally, it mentions specific tools for data analysis and retrieval, further highlighting the critical role databases play in the field of molecular biology.