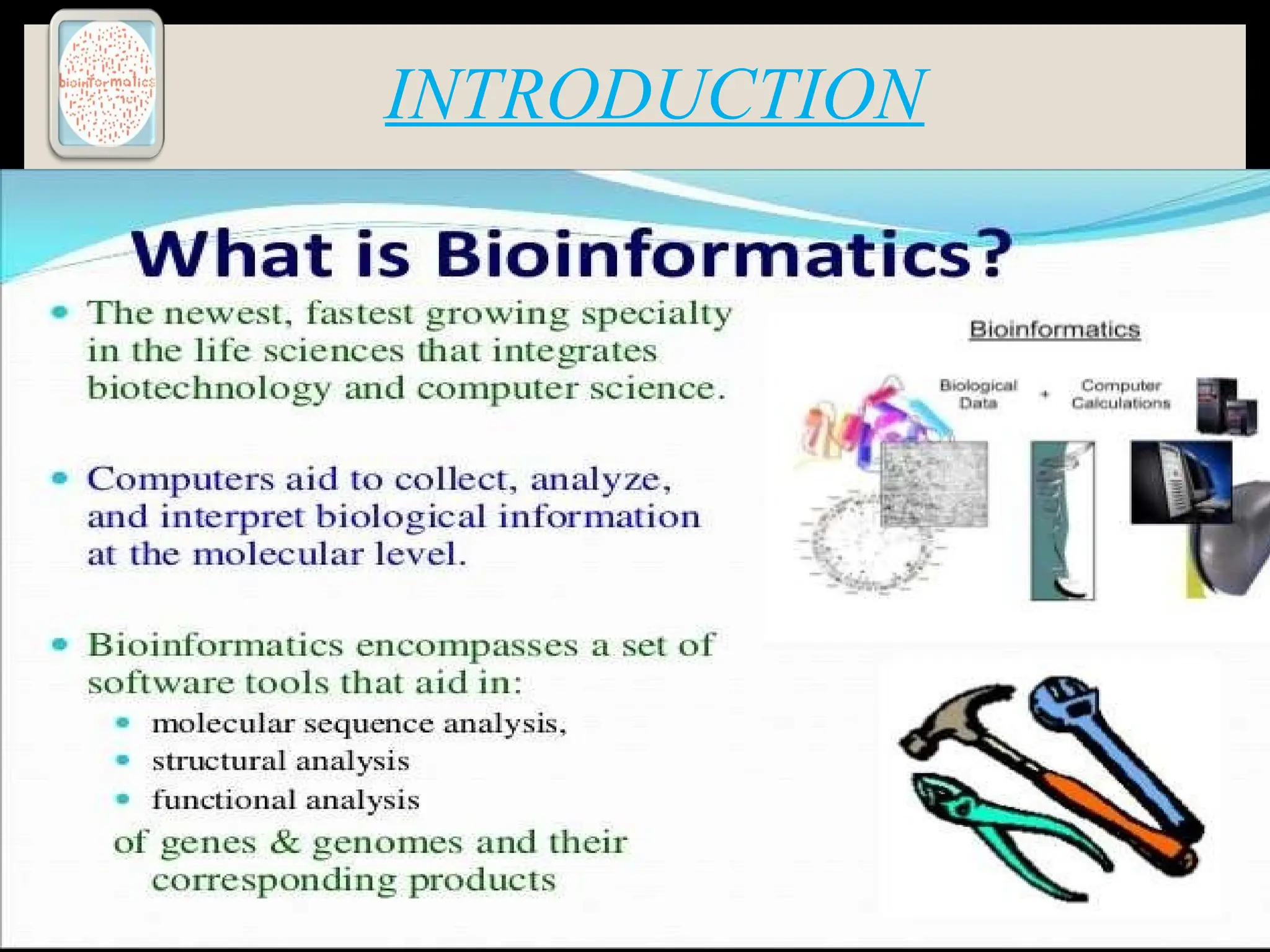
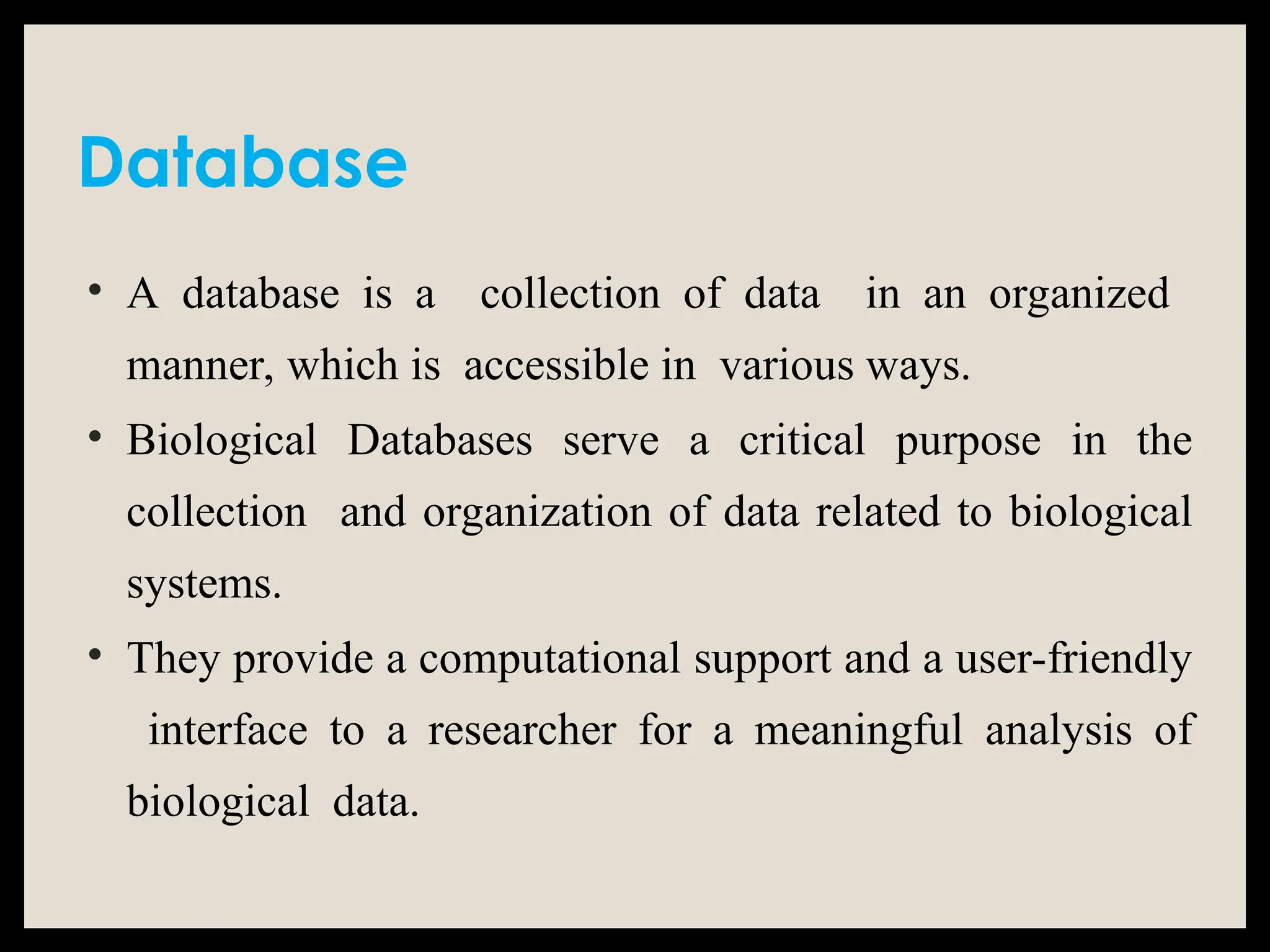
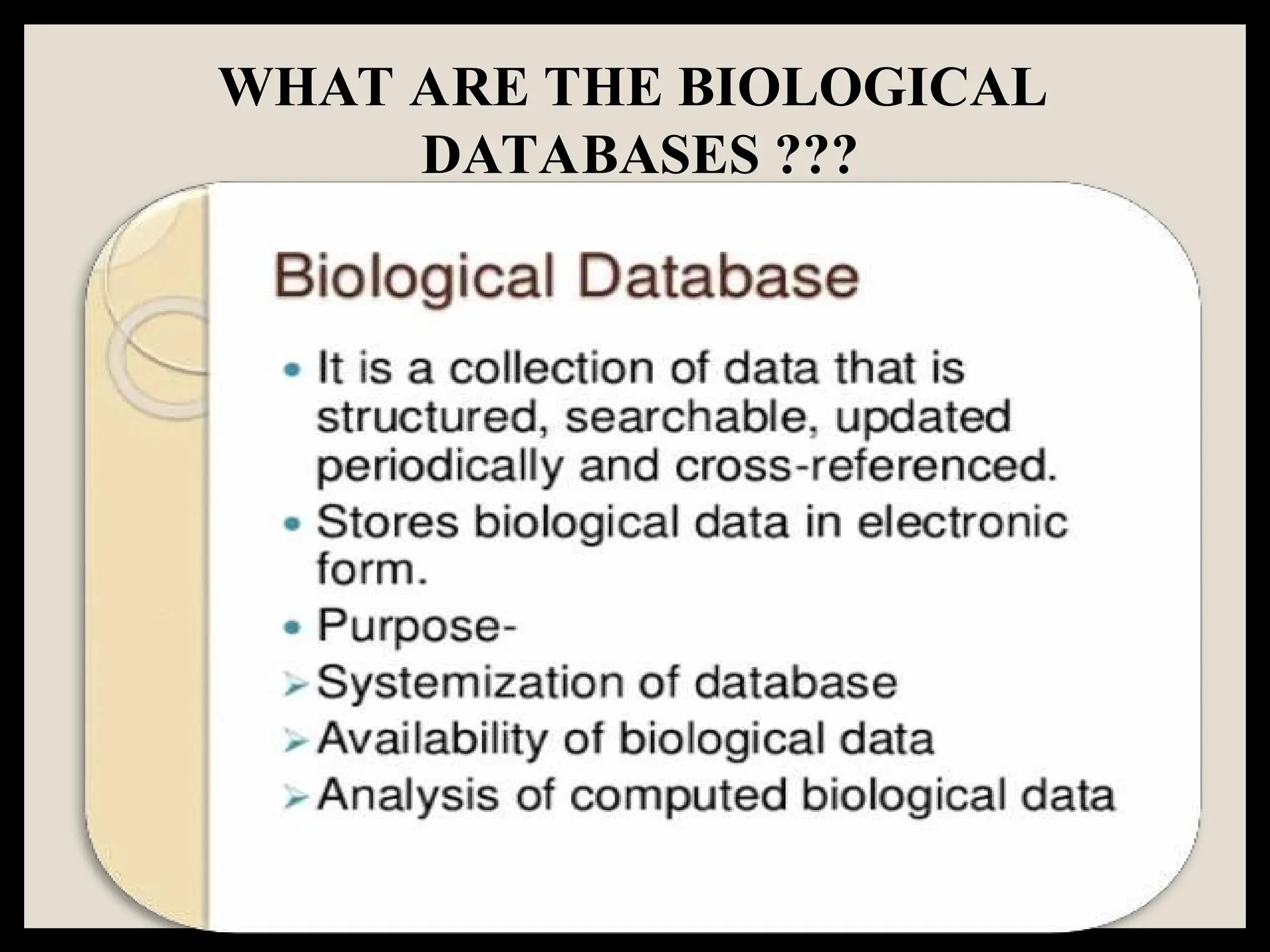
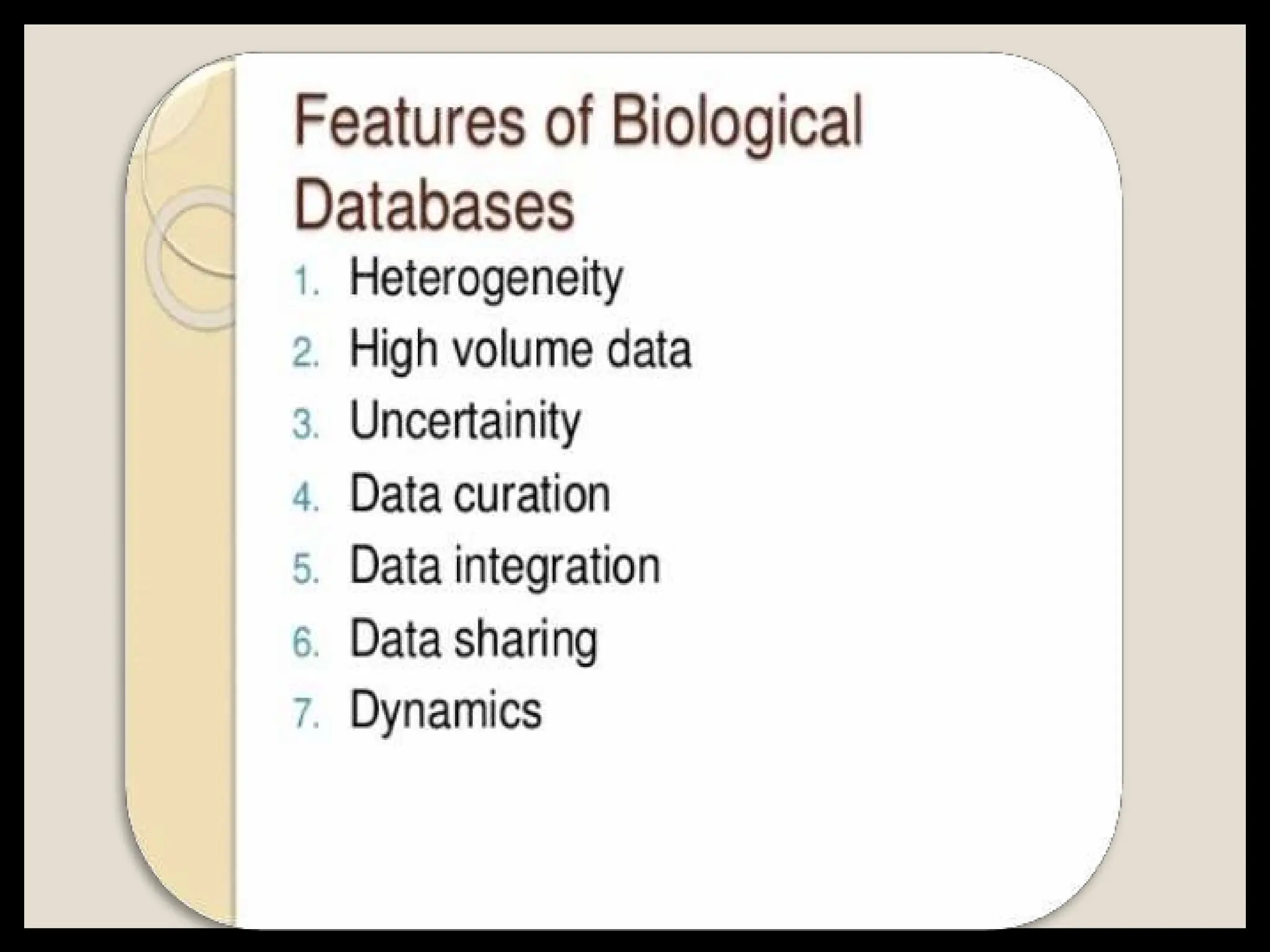
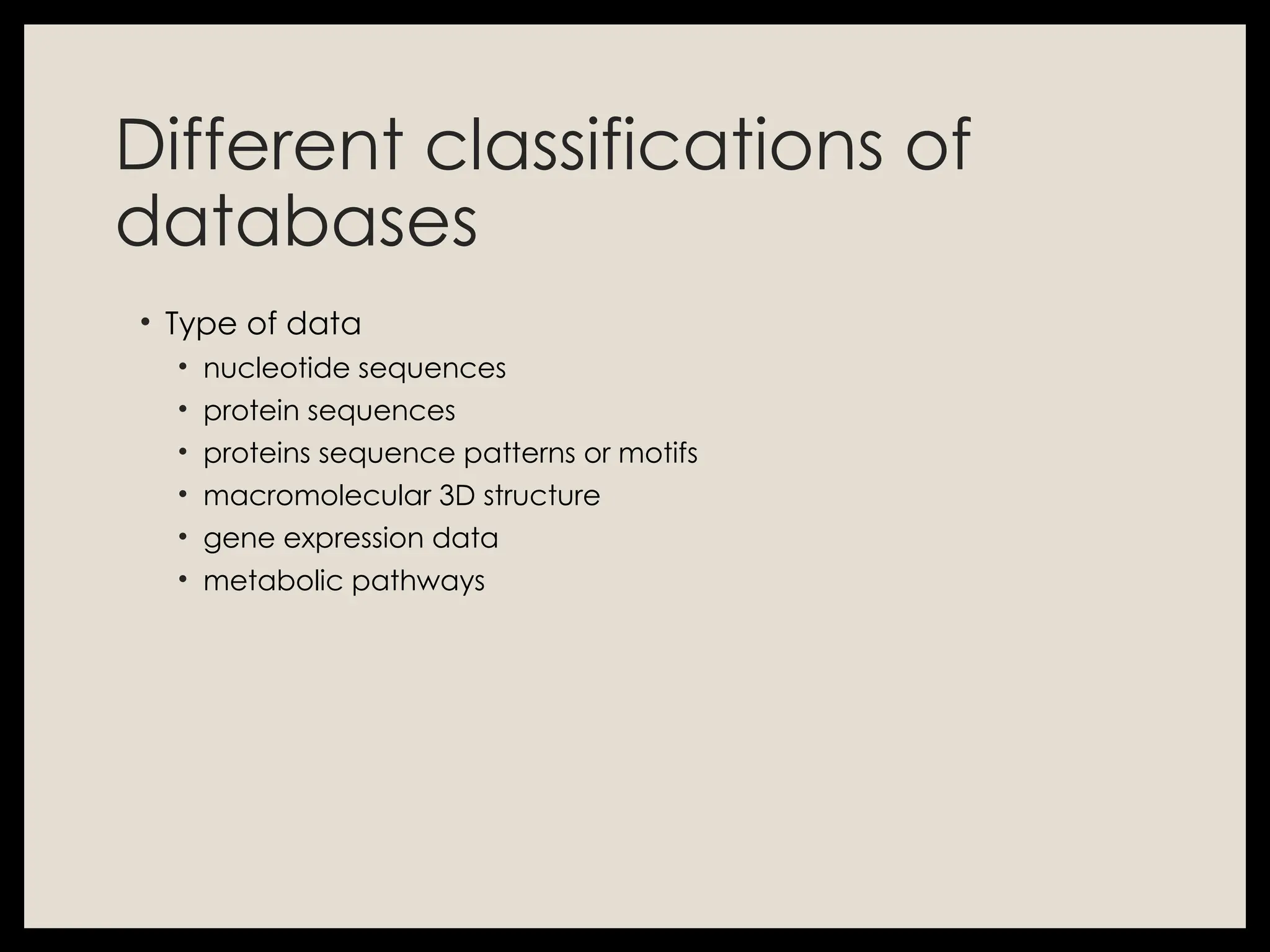
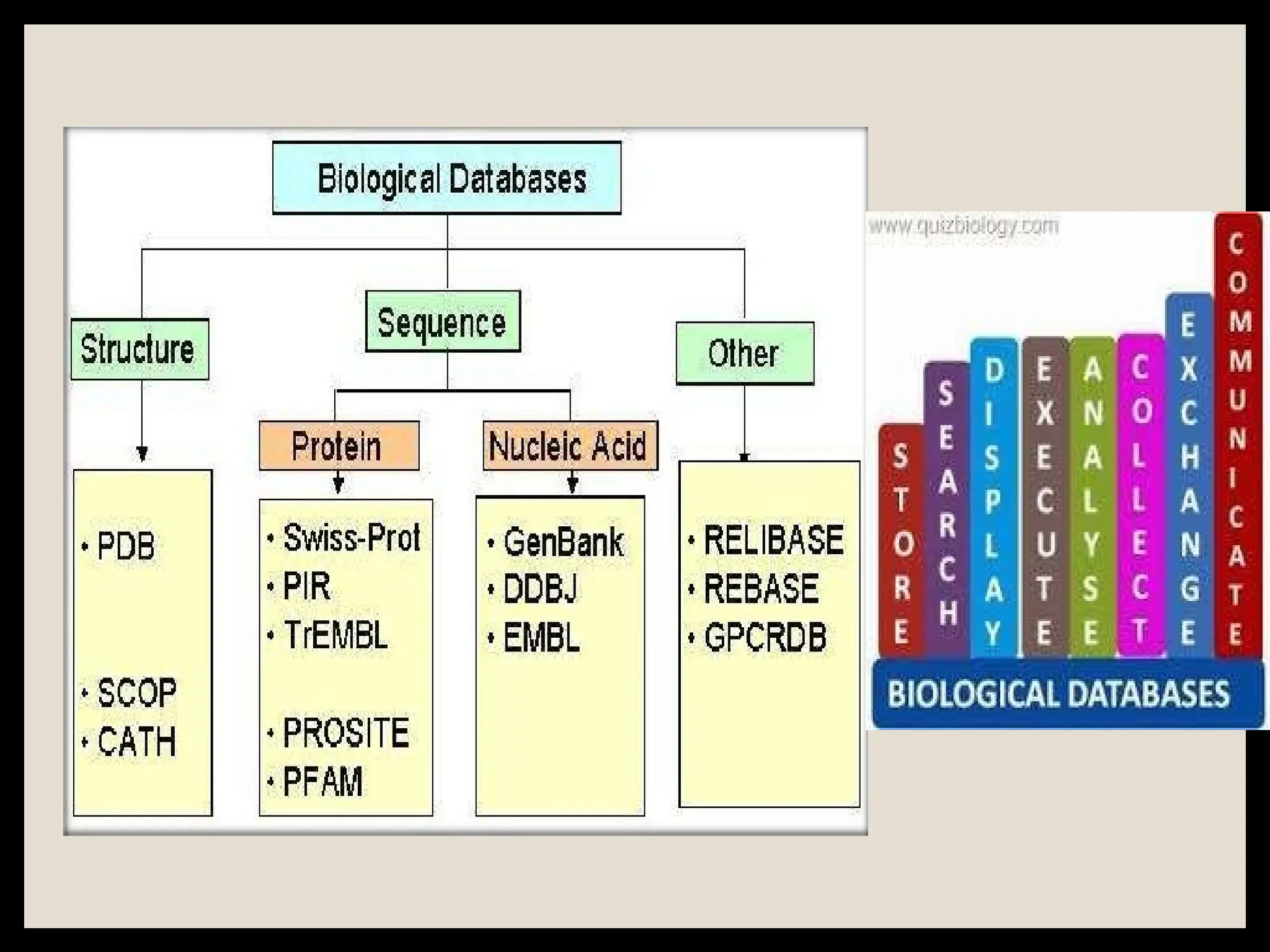
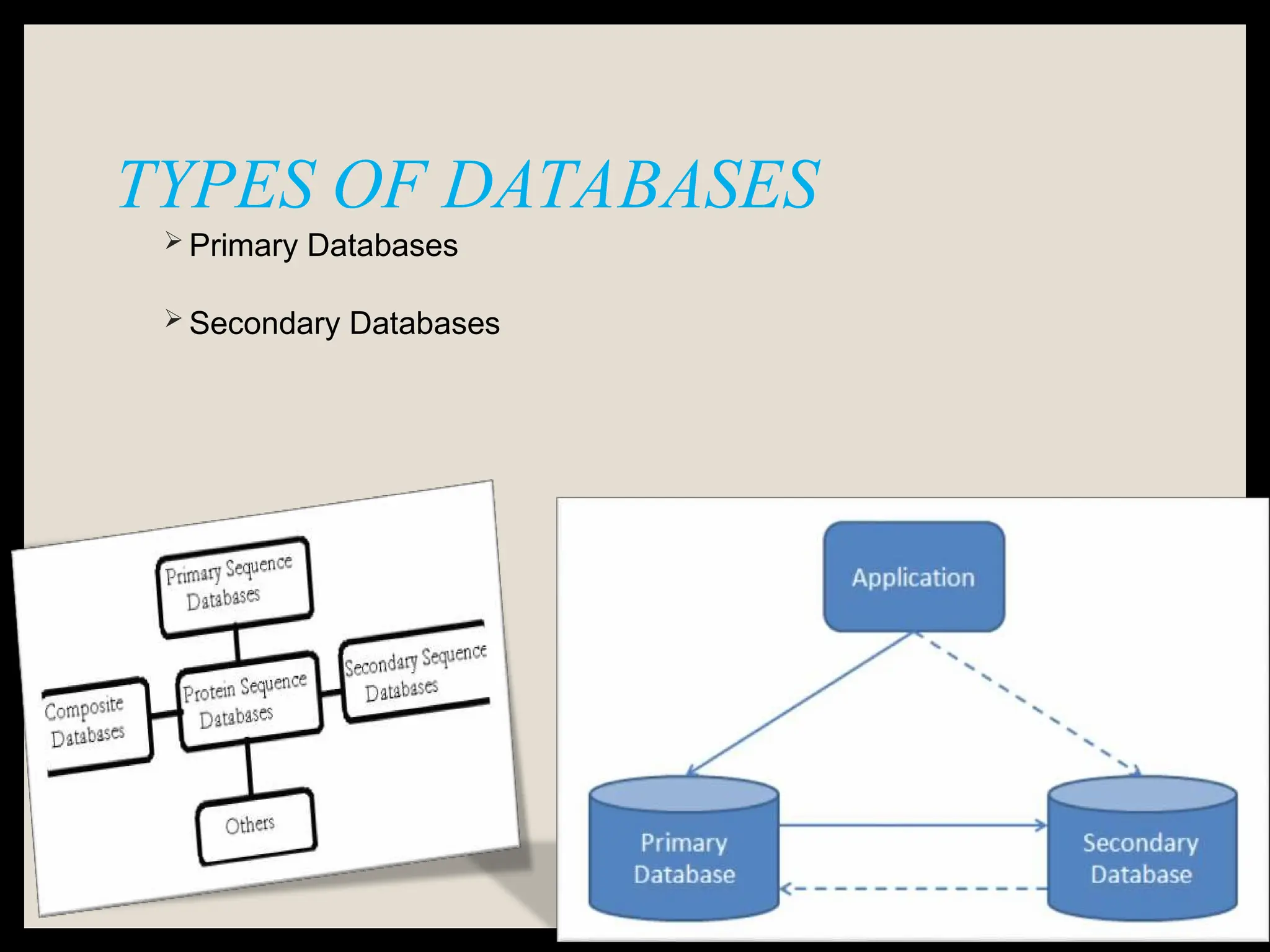
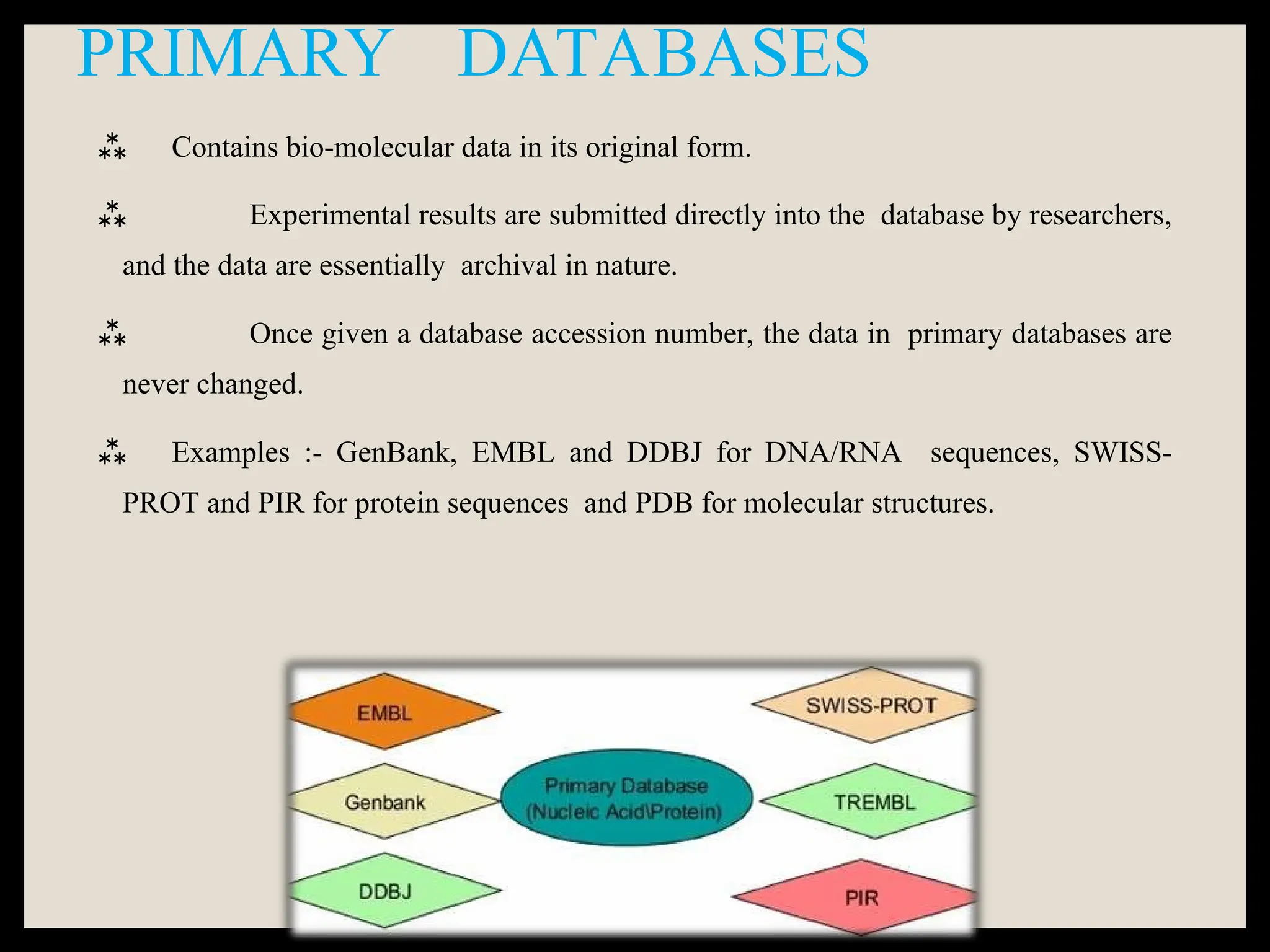
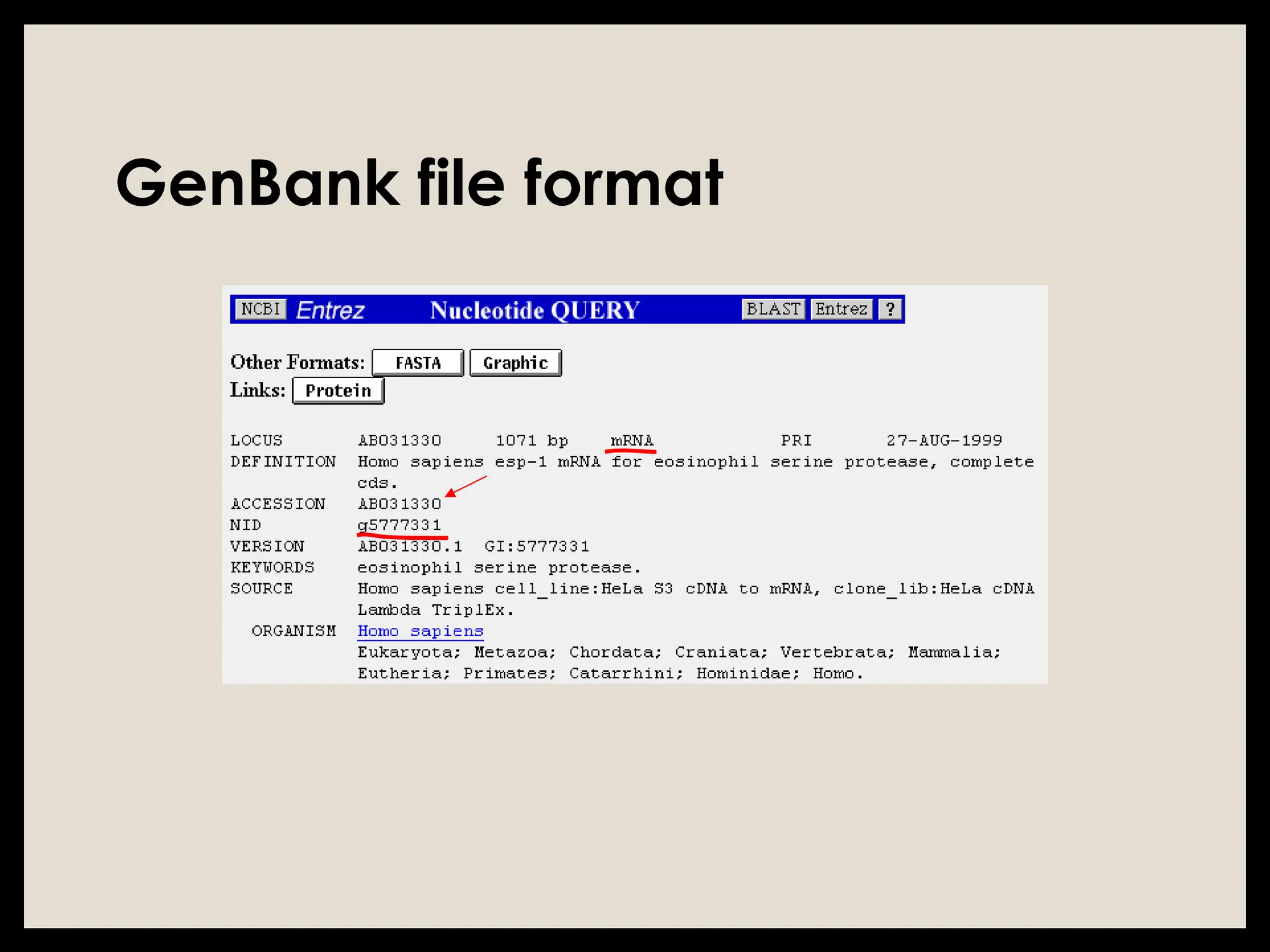
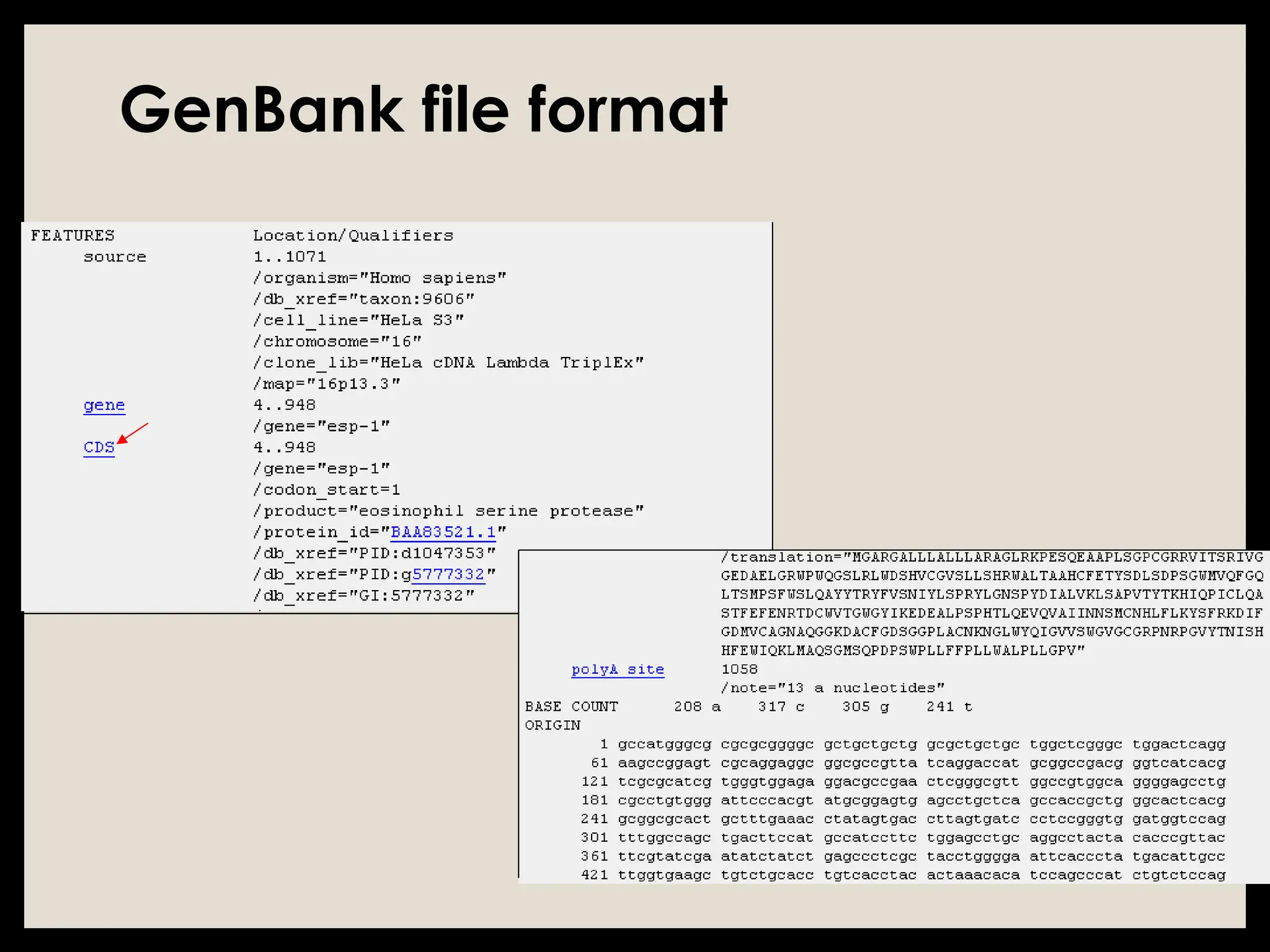
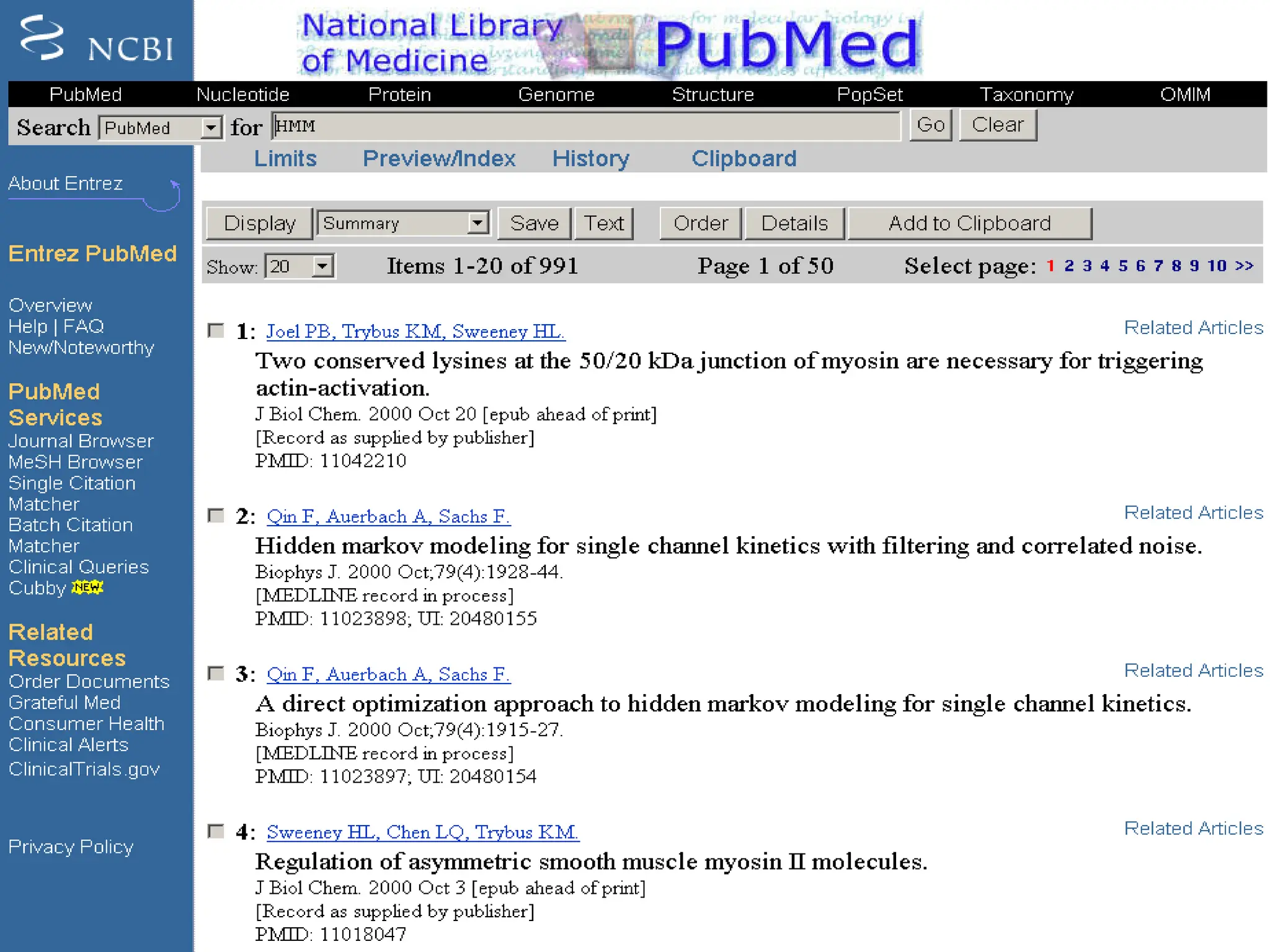
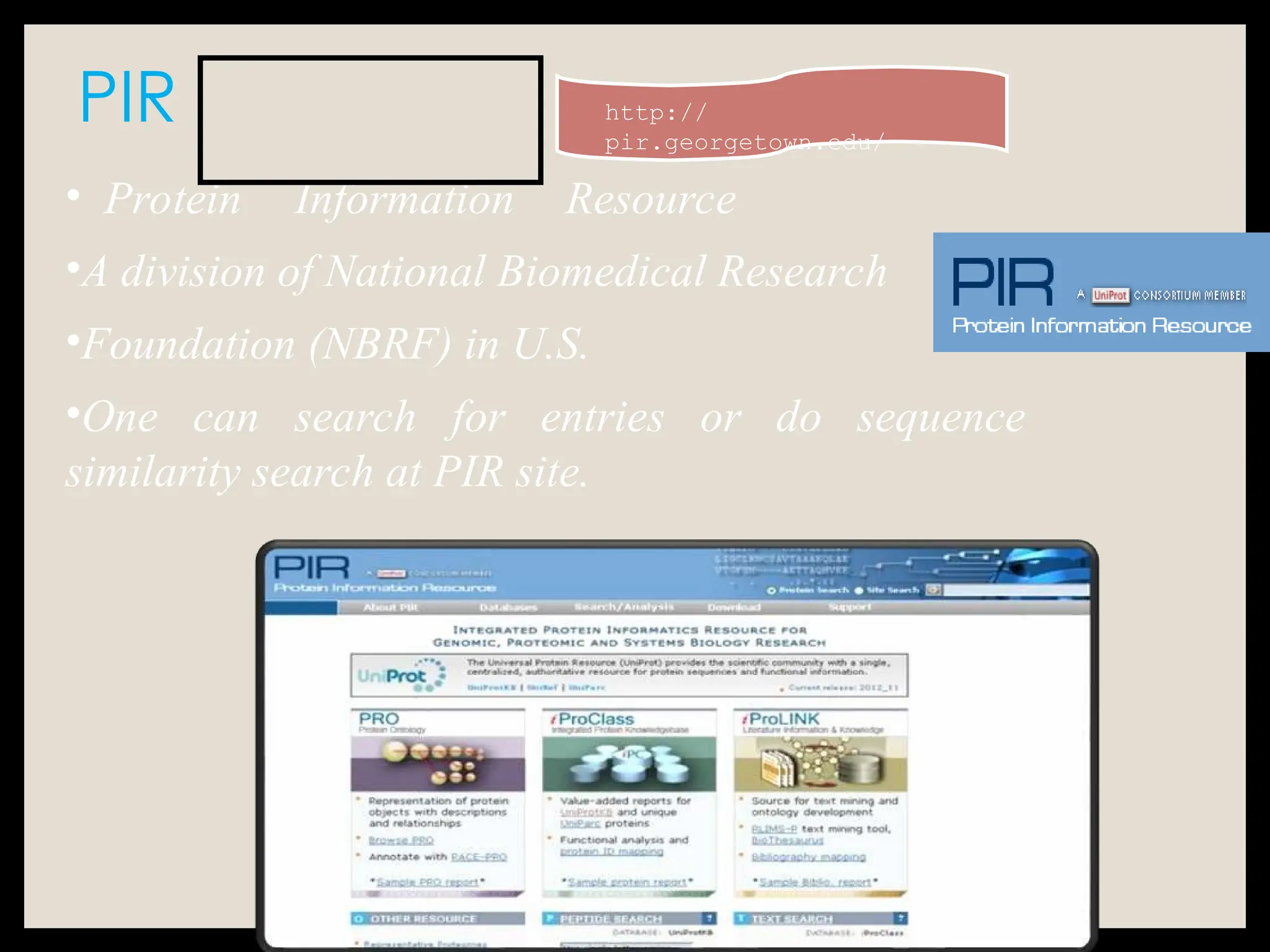
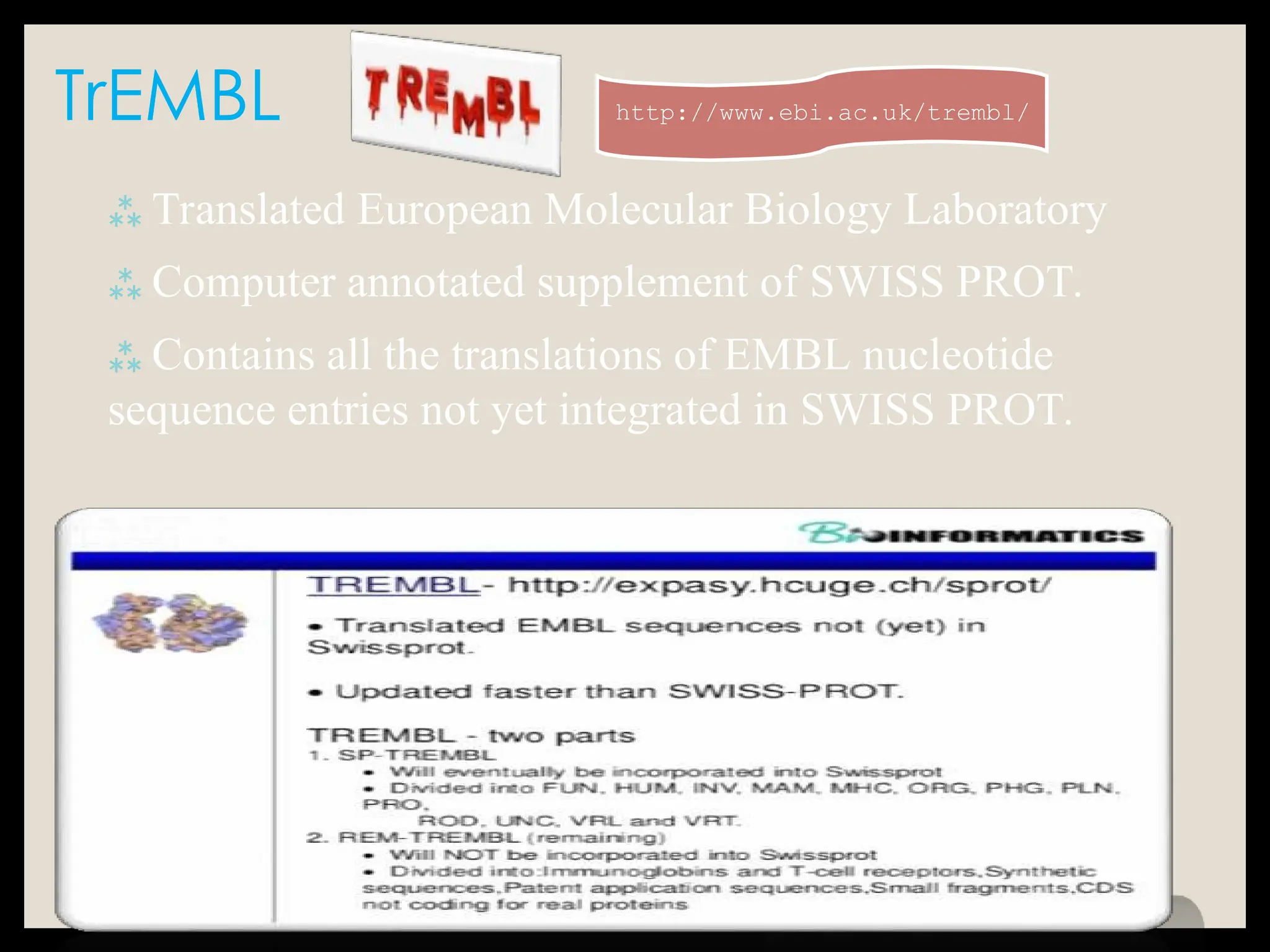
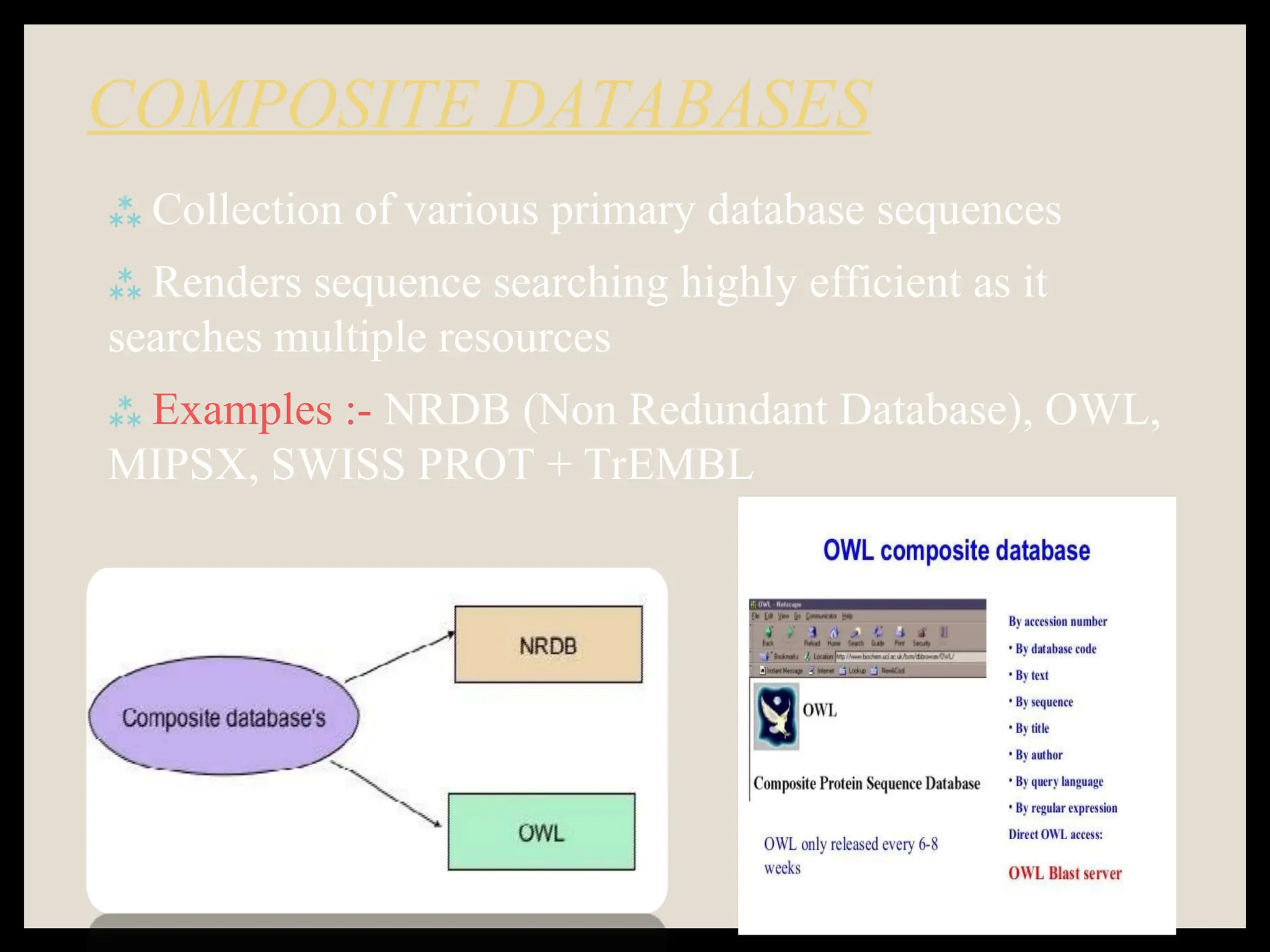
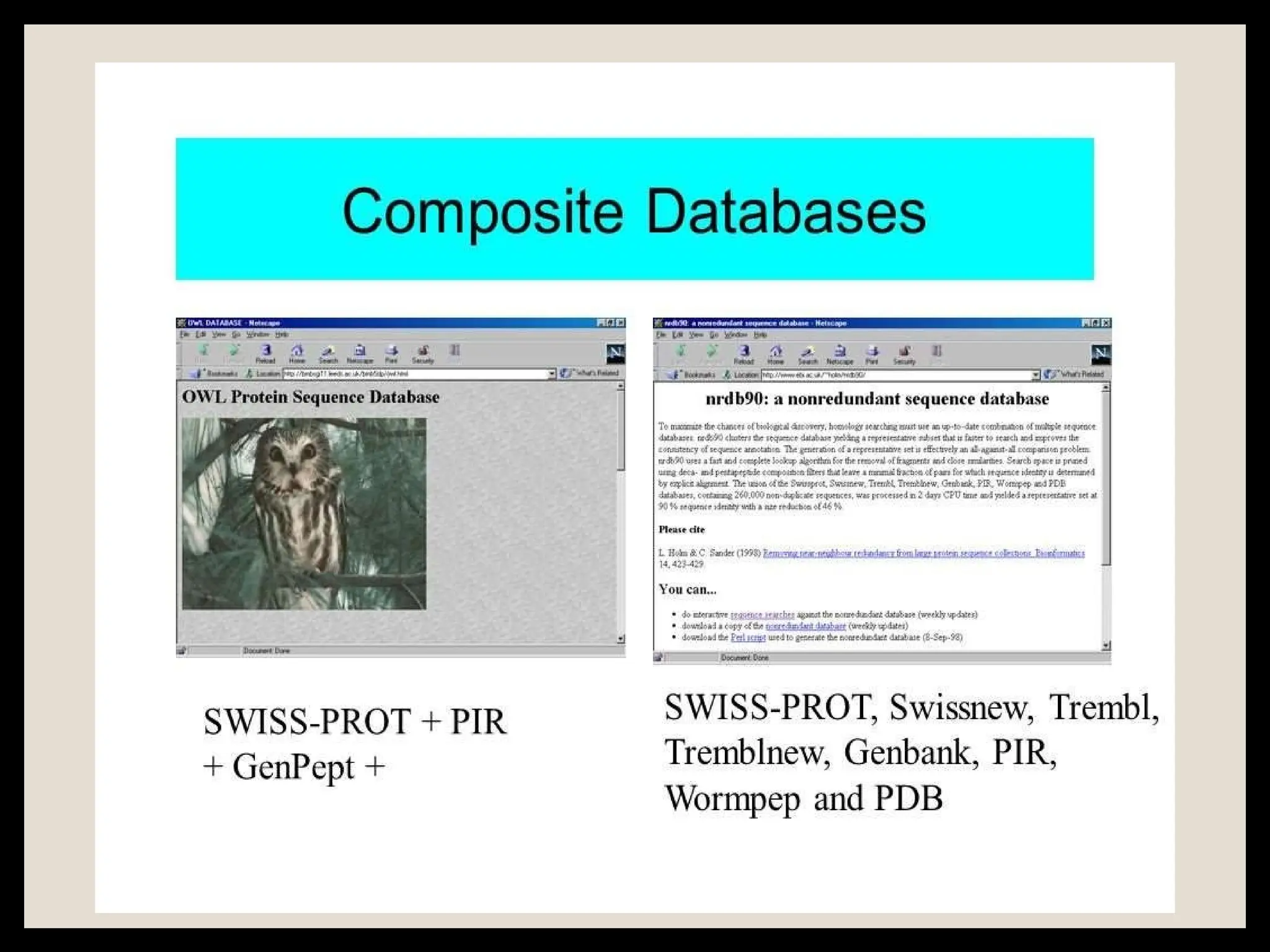
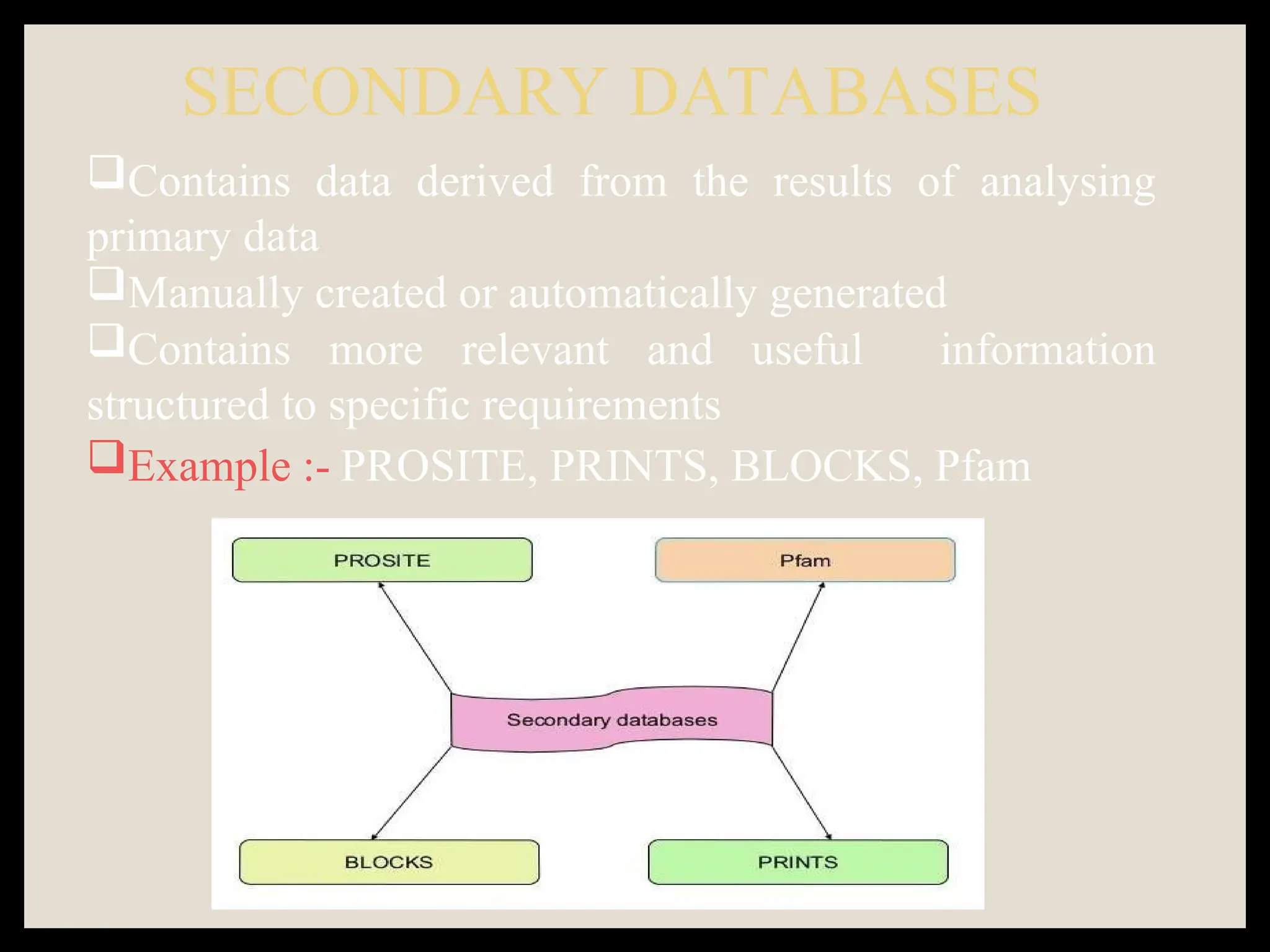
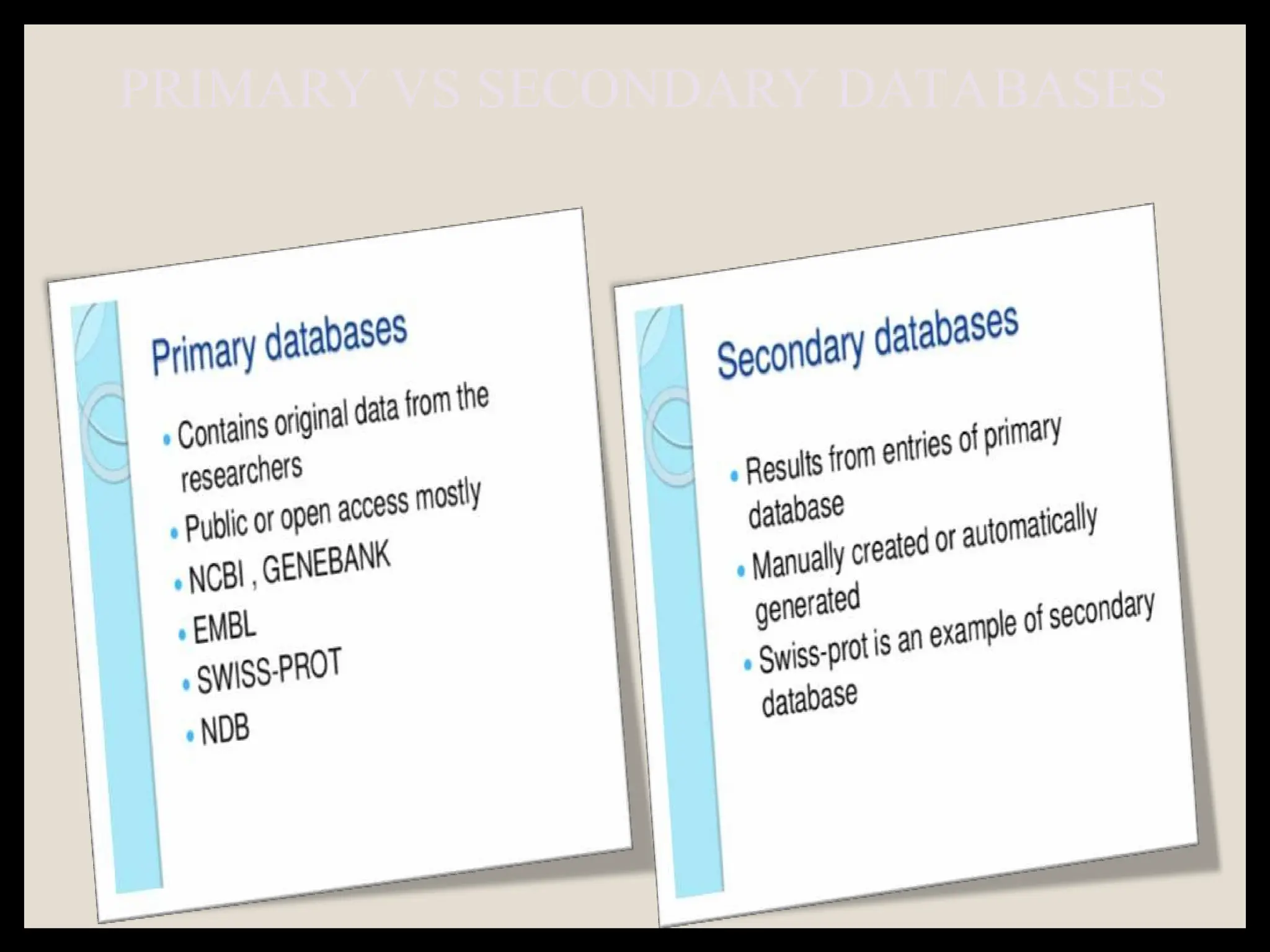
The document provides an overview of databases, differentiating between raw data and processed information. It explains various types of biological databases, including primary and secondary databases, and their classifications based on data types and availability. Examples of significant databases such as GenBank, EMBL, and PDB are also discussed, highlighting their roles in biological research and data management.