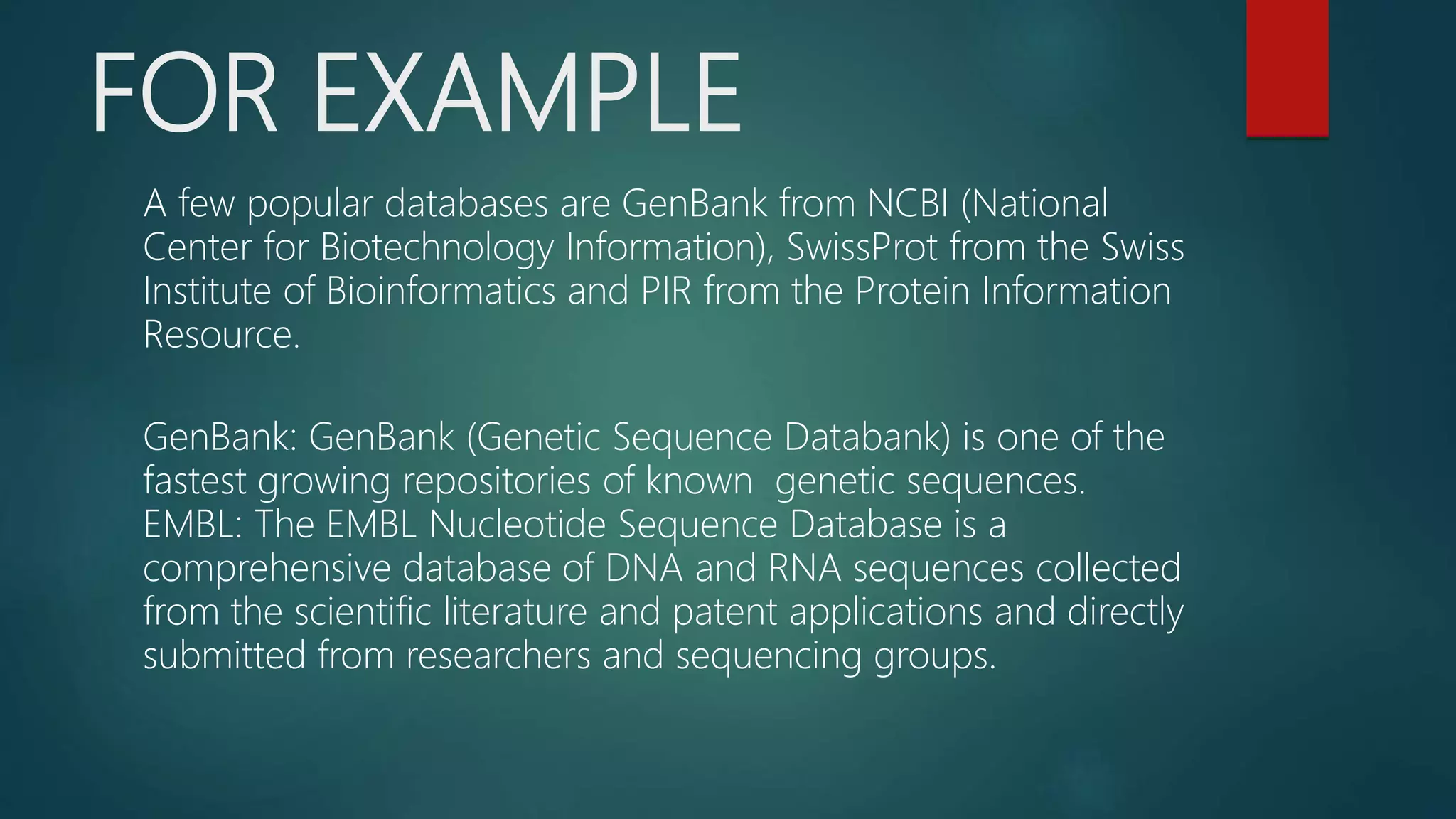
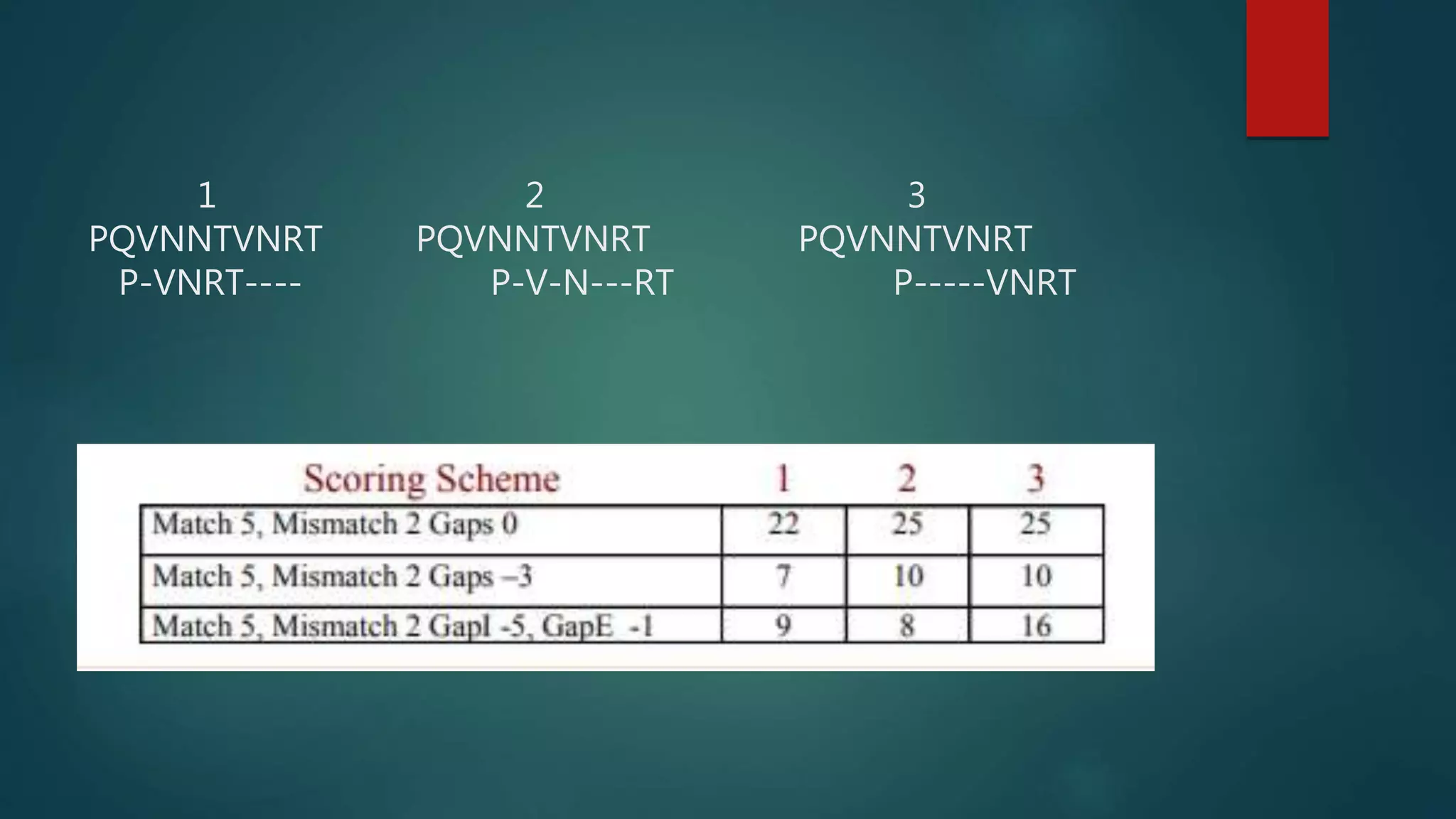
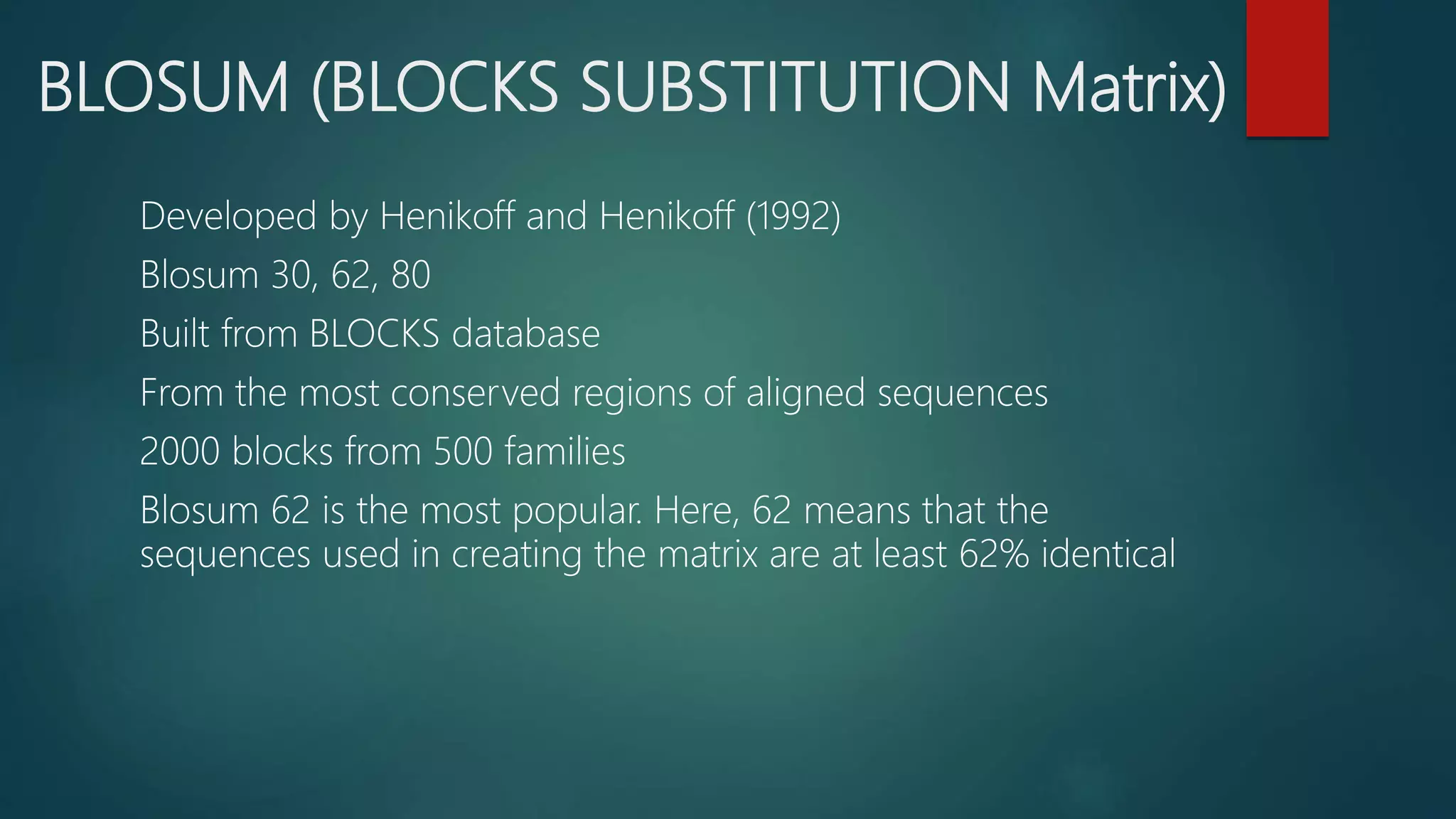
This document discusses biological database searching. It begins by defining biological databases as organized collections of persistent biological data that can be queried and retrieved. Examples are provided such as GenBank and SwissProt. Database searching involves using a query sequence to find related sequences in the database based on homology. Parameters like E-values and bit scores are used to define the significance of matches. Popular search algorithms like BLAST and FASTA first use heuristics to find high-scoring regions before applying dynamic programming for alignment.