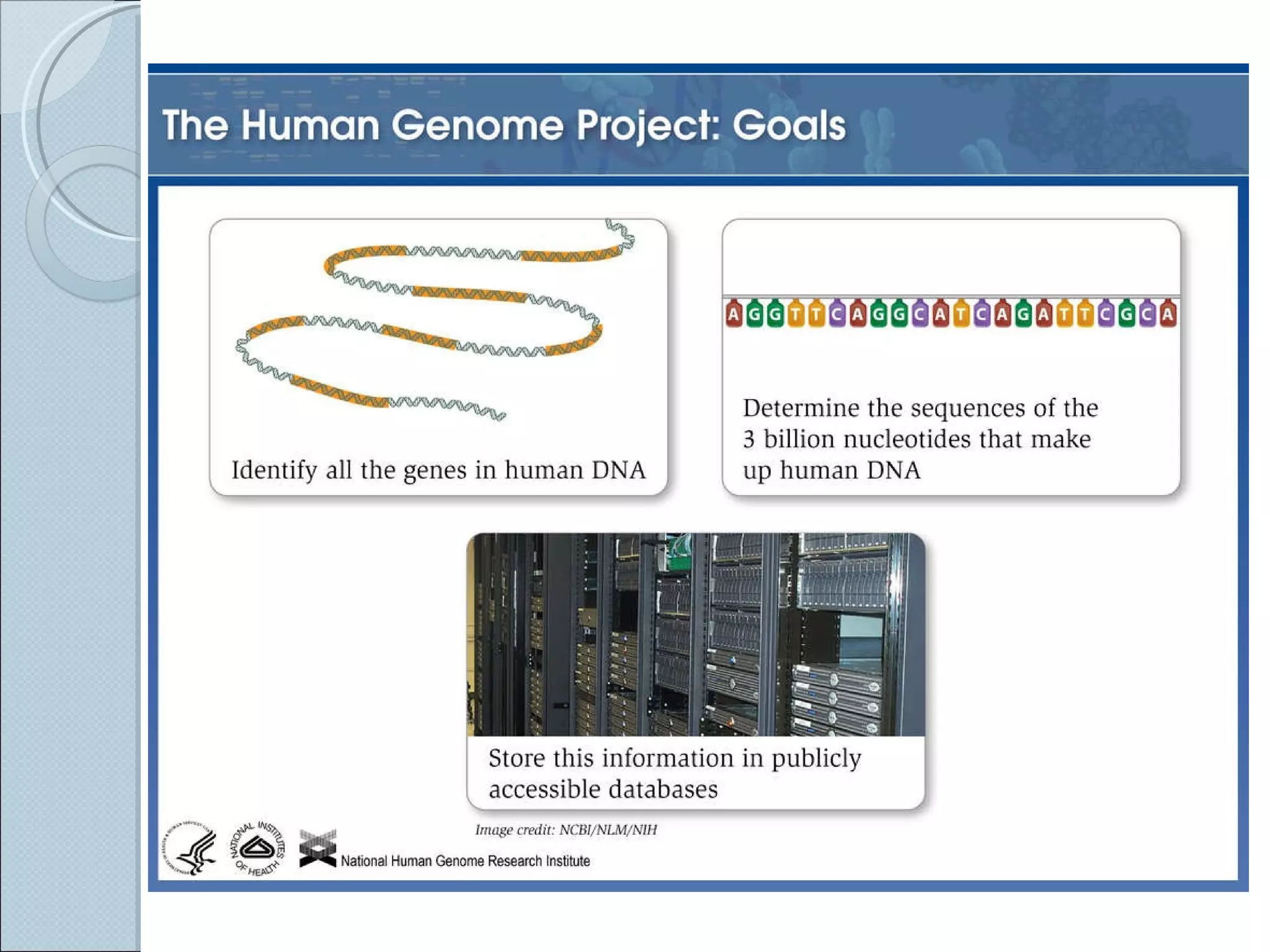
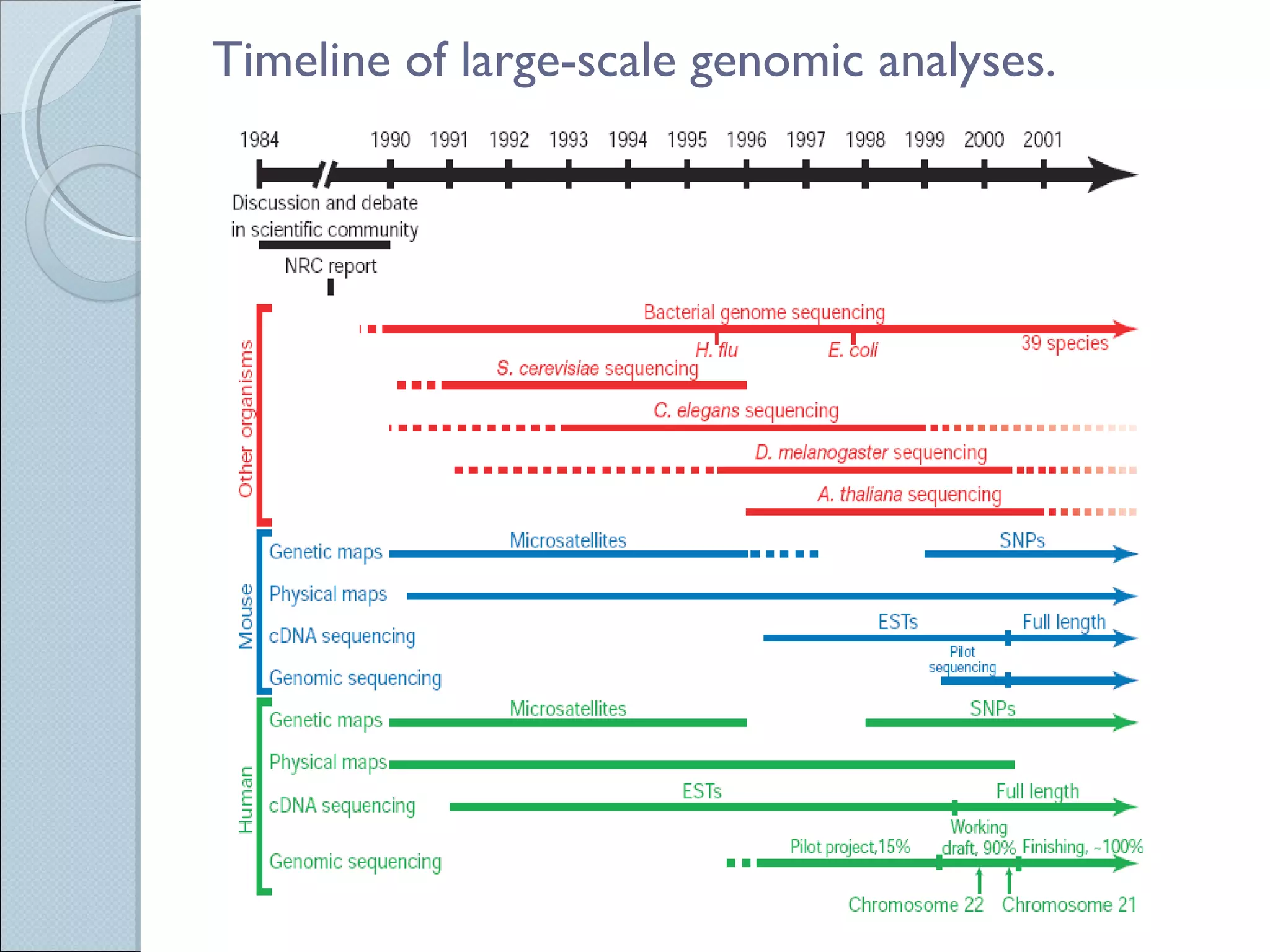
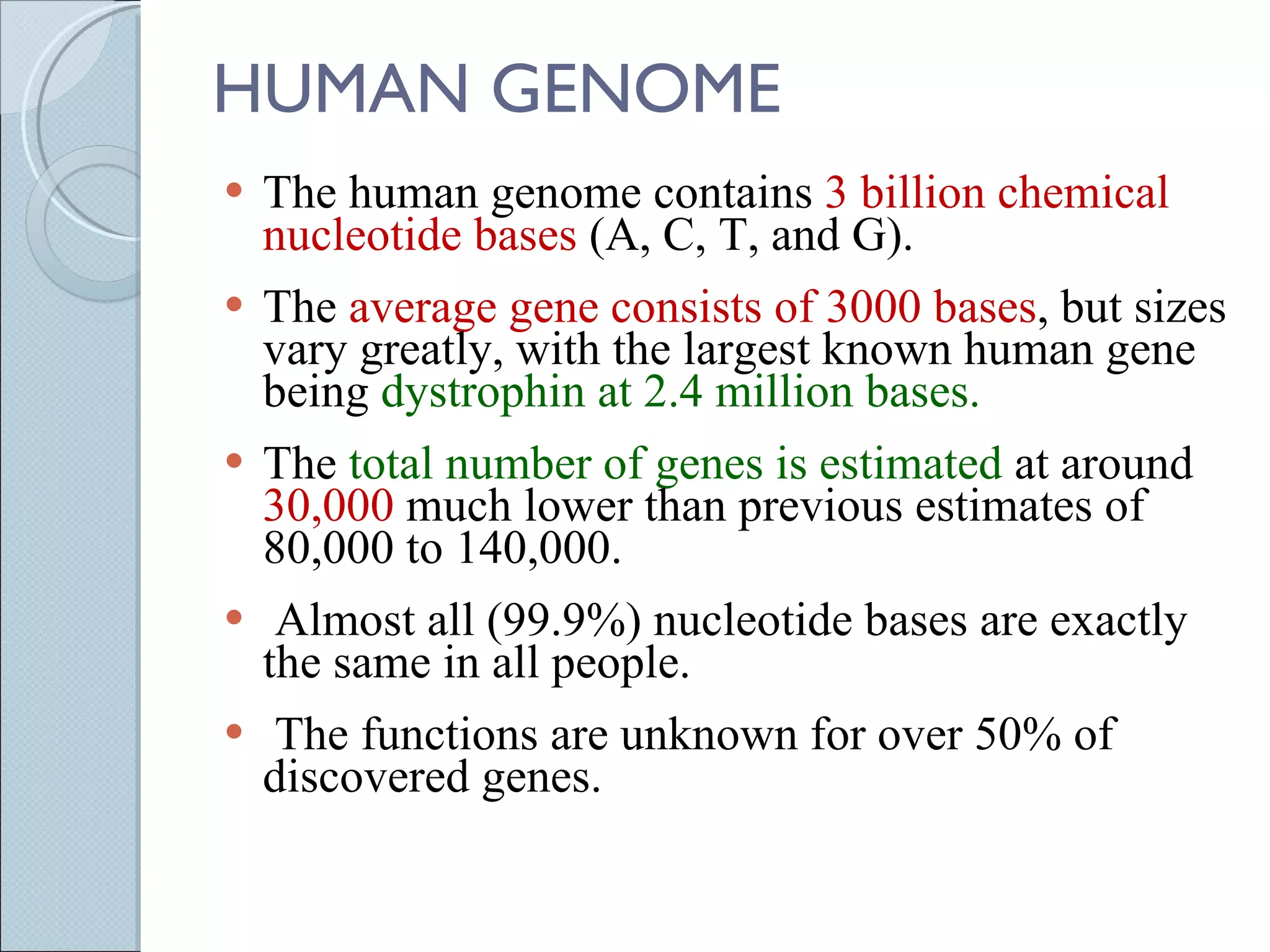
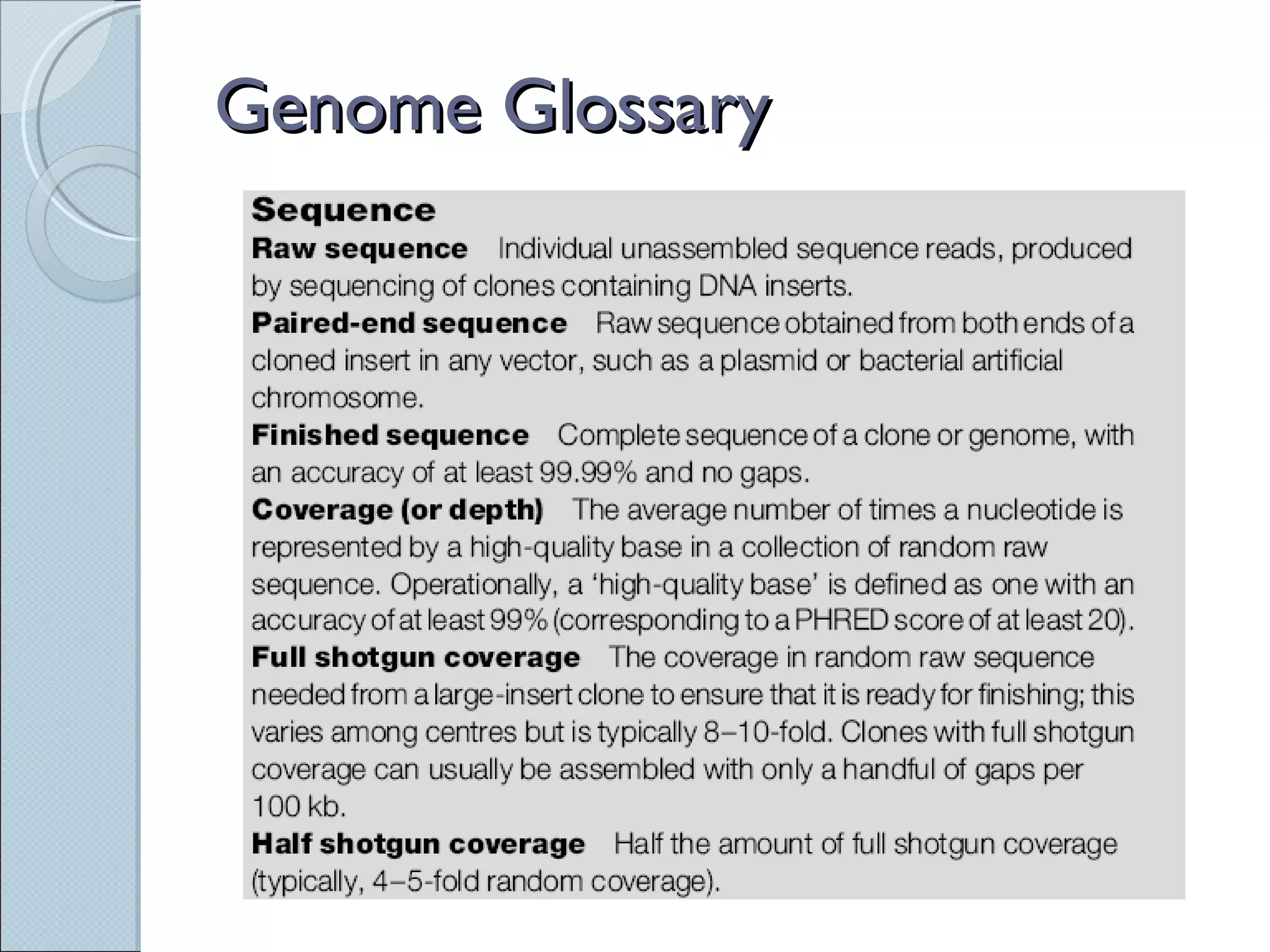
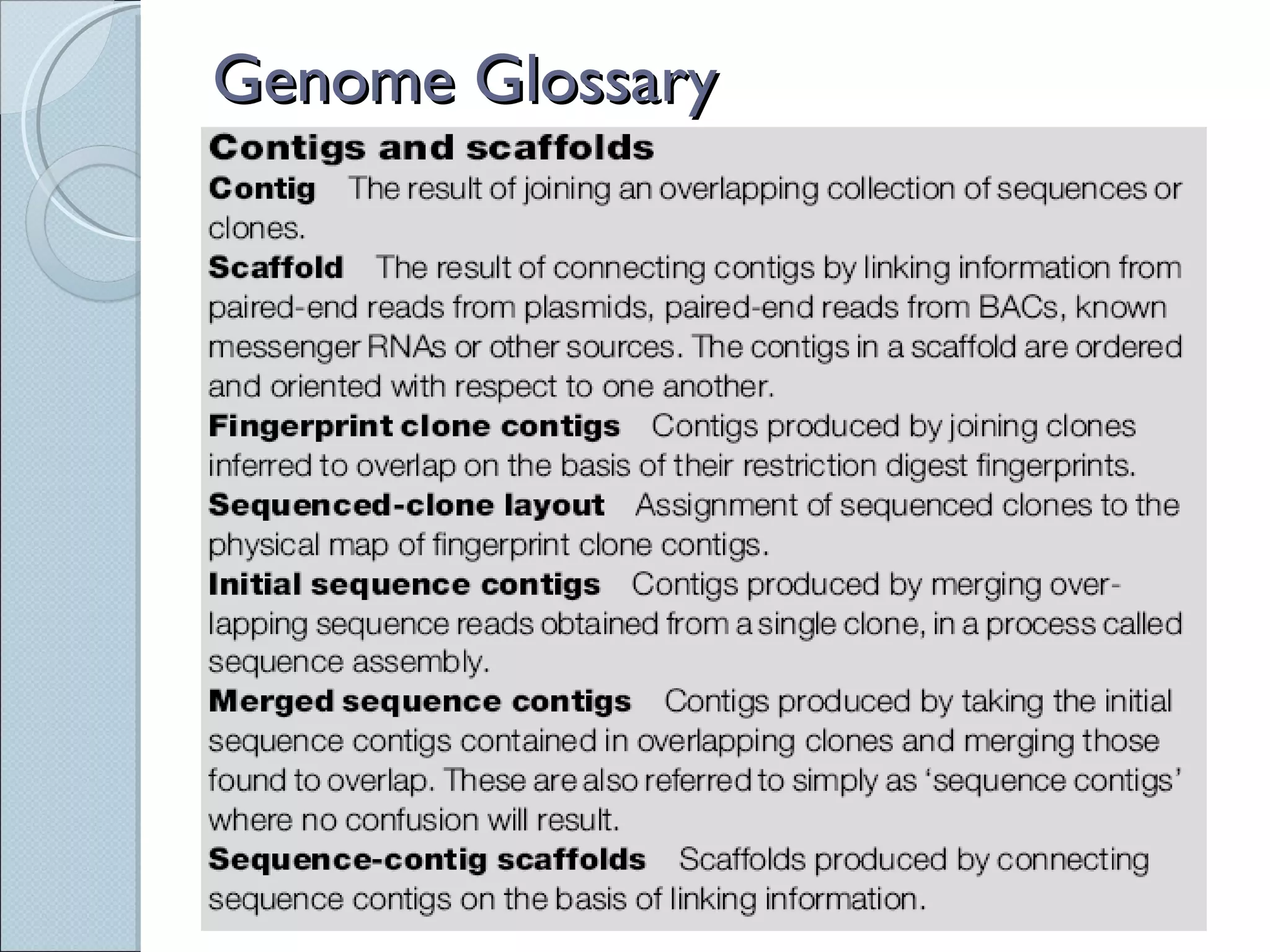
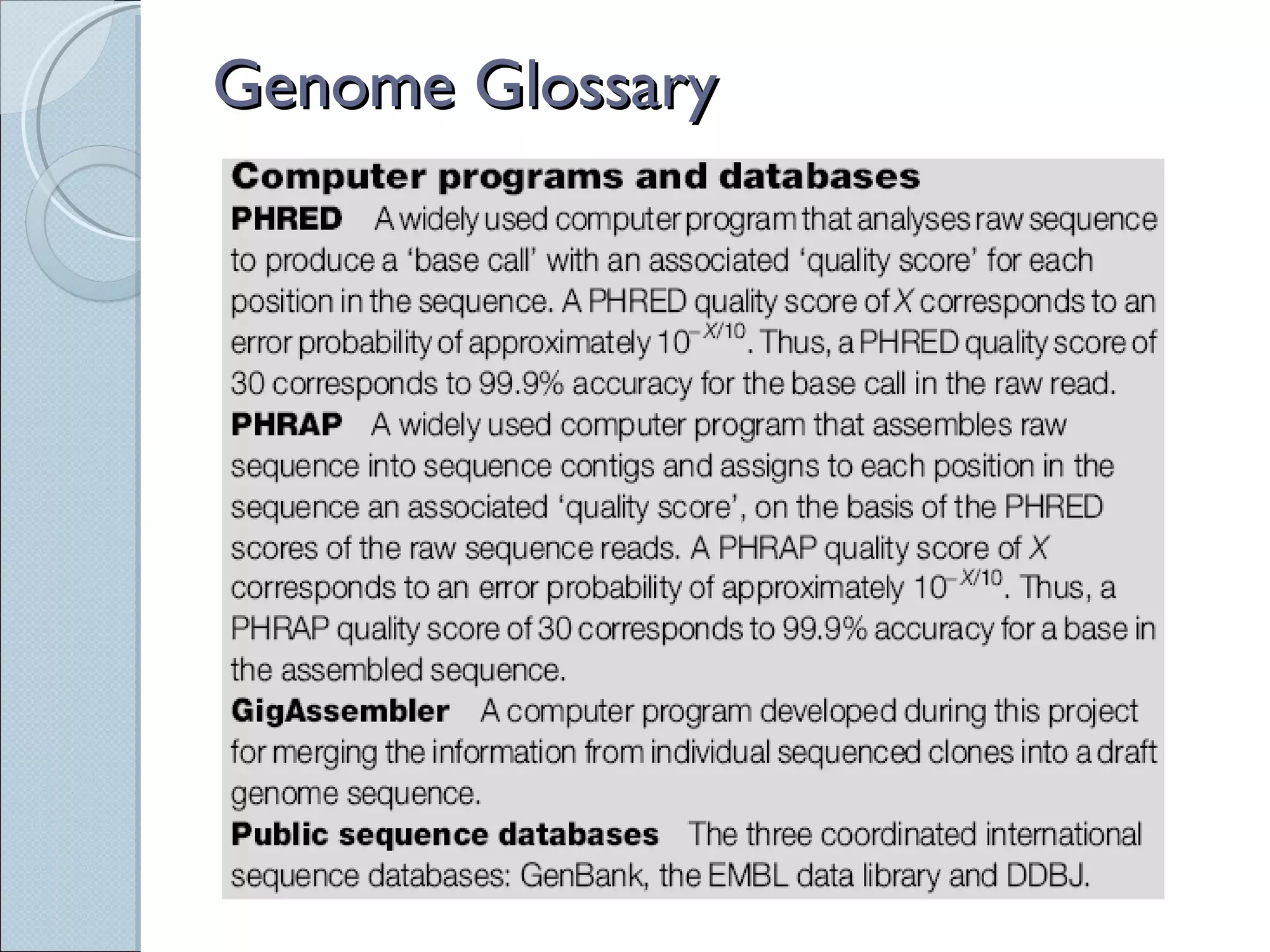
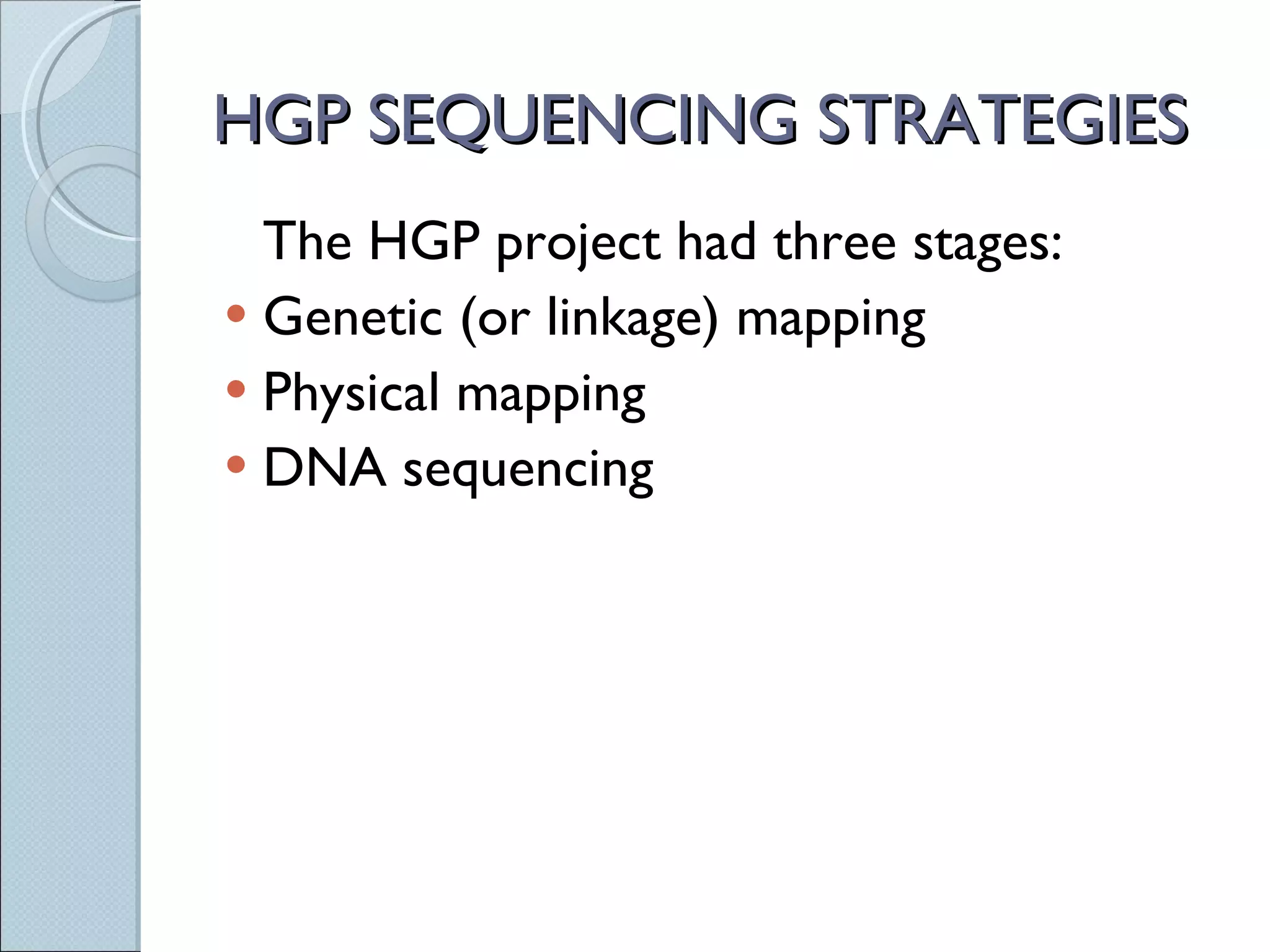
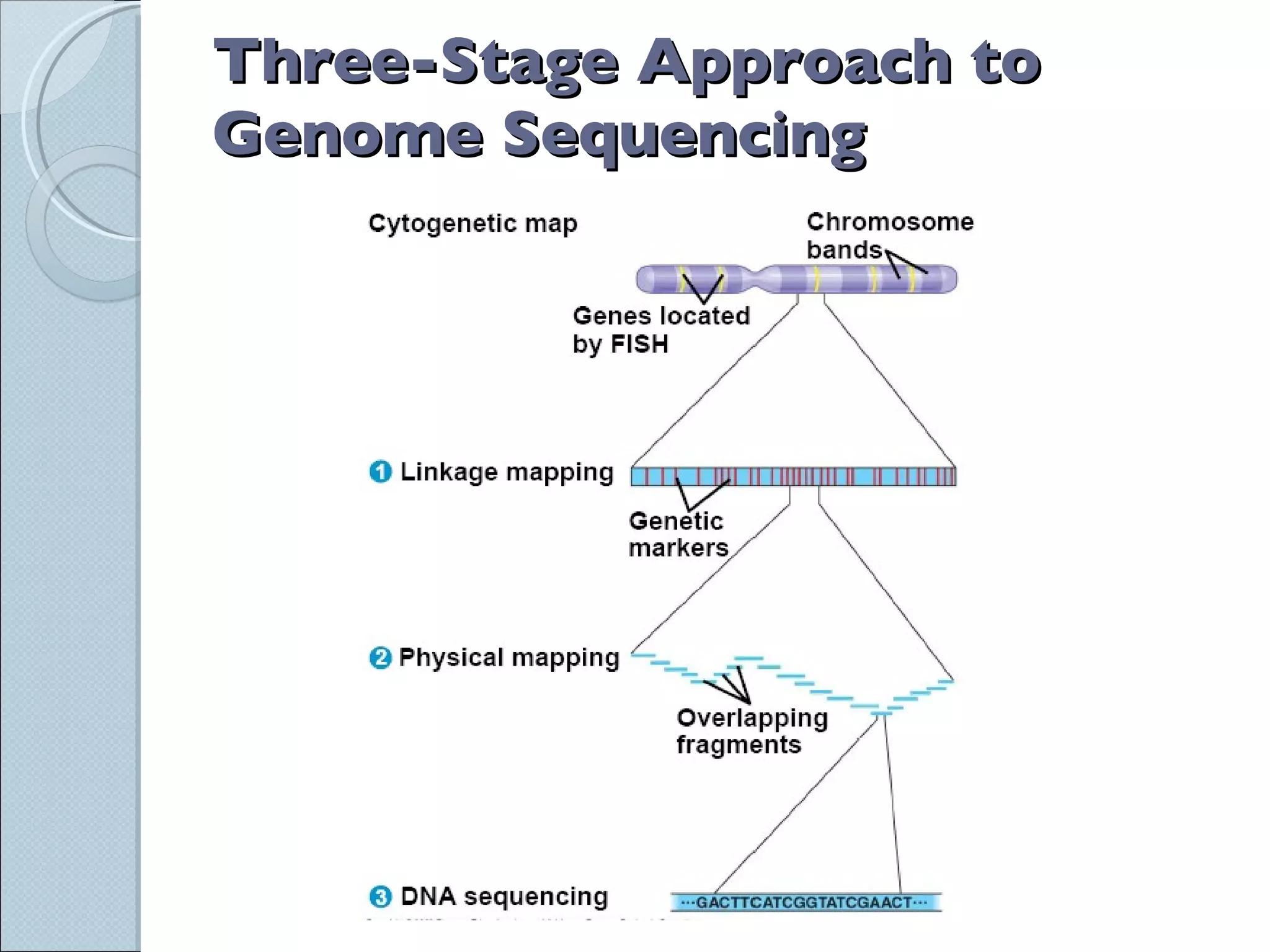
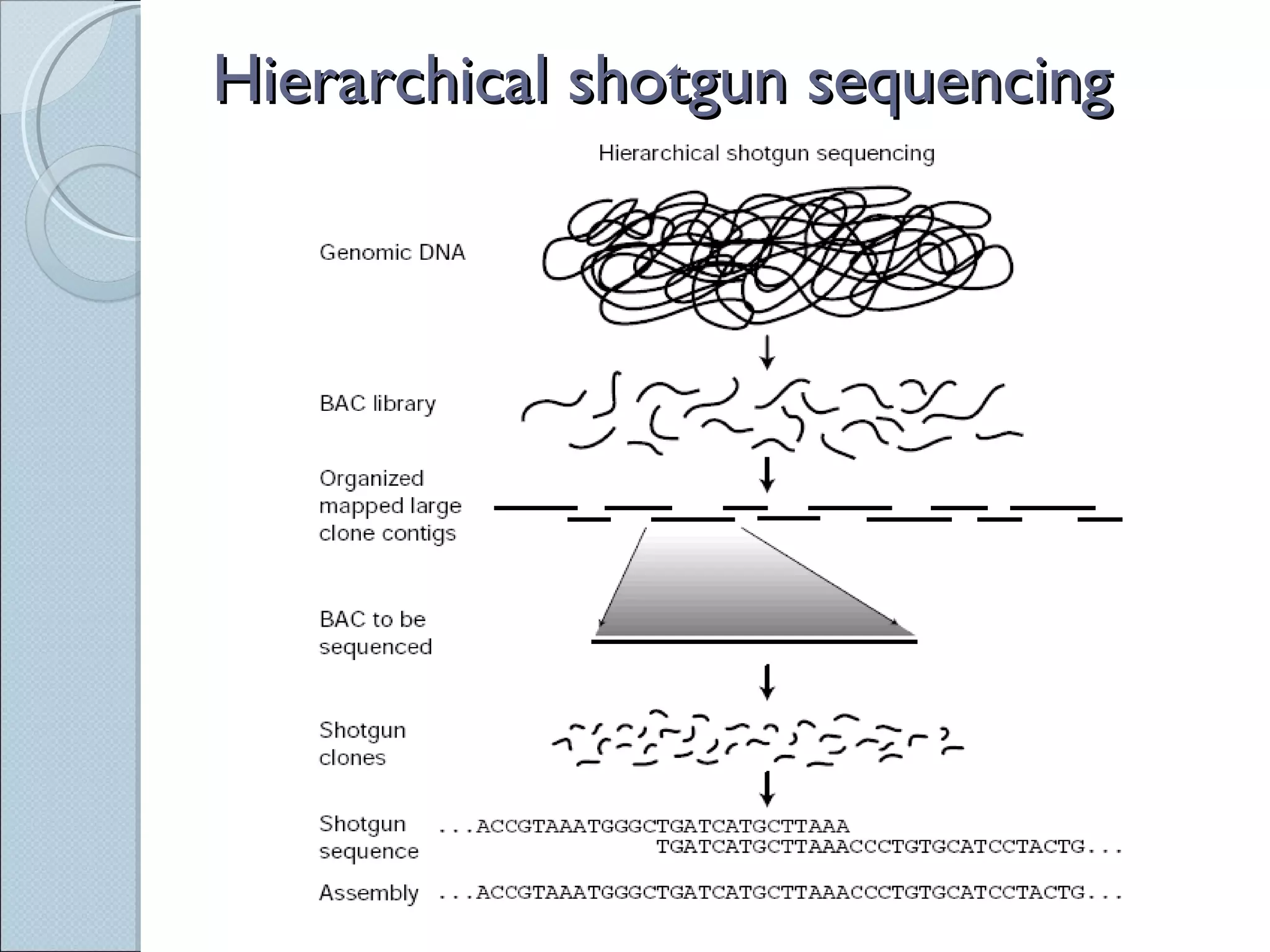
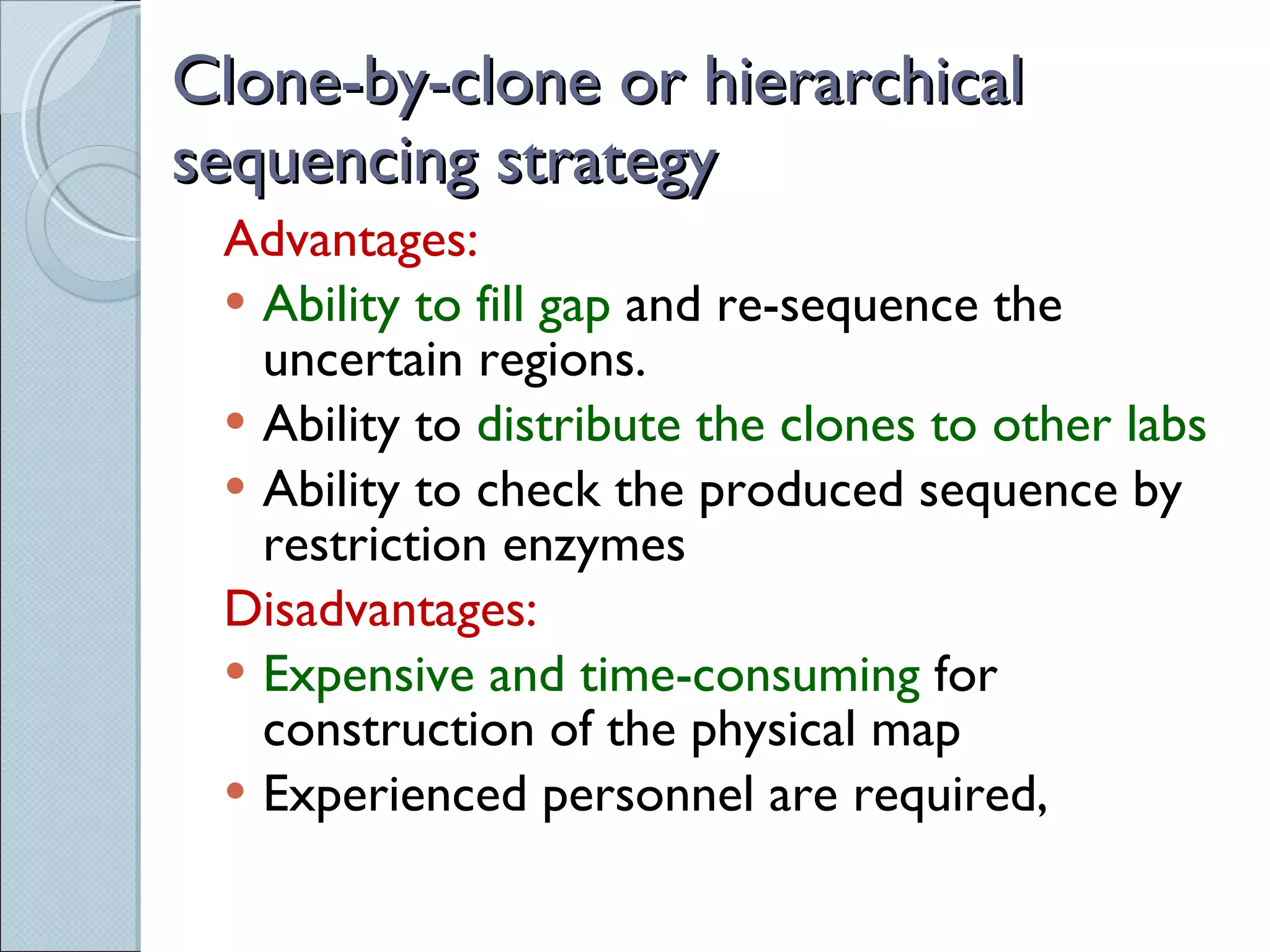
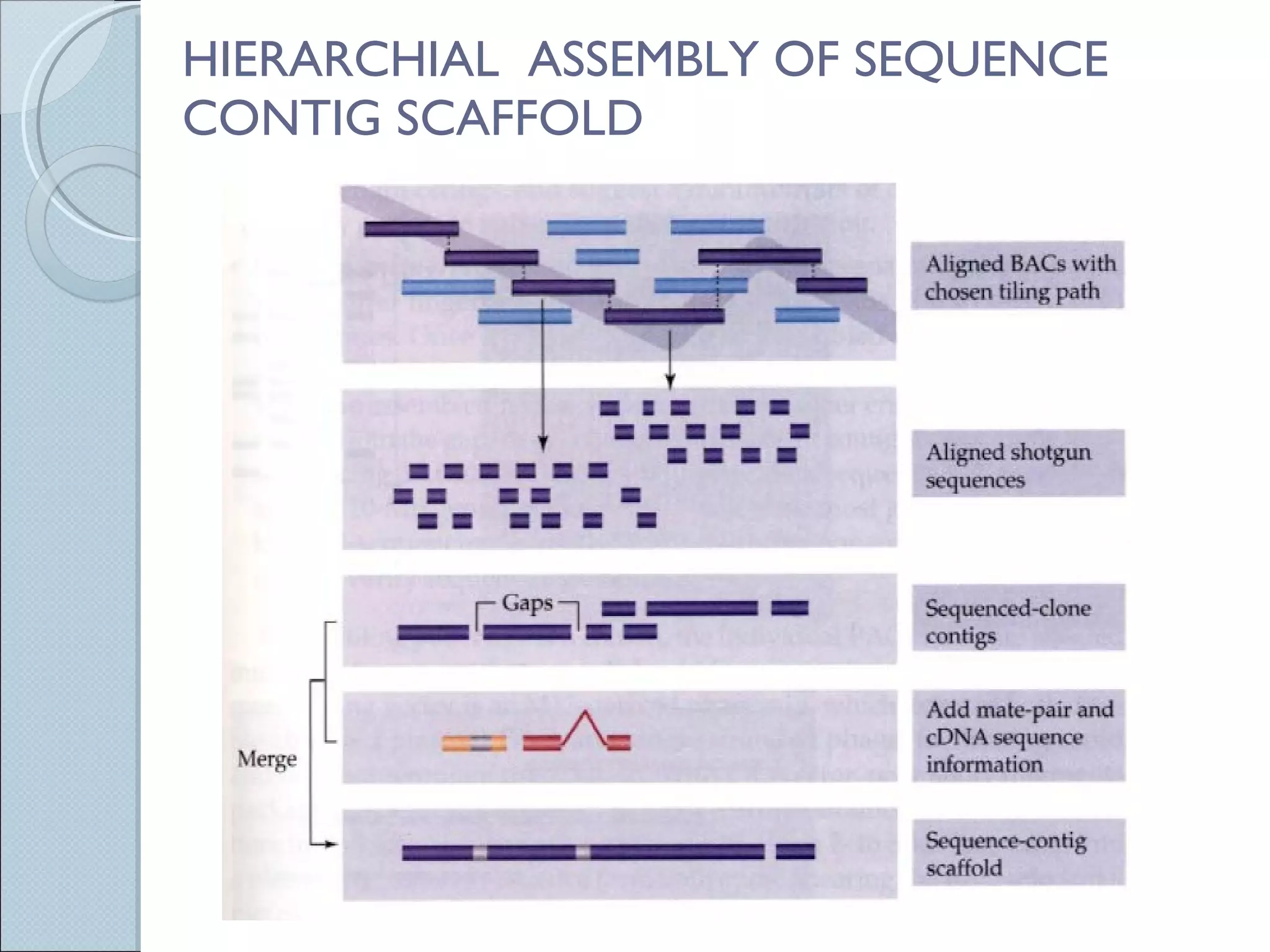
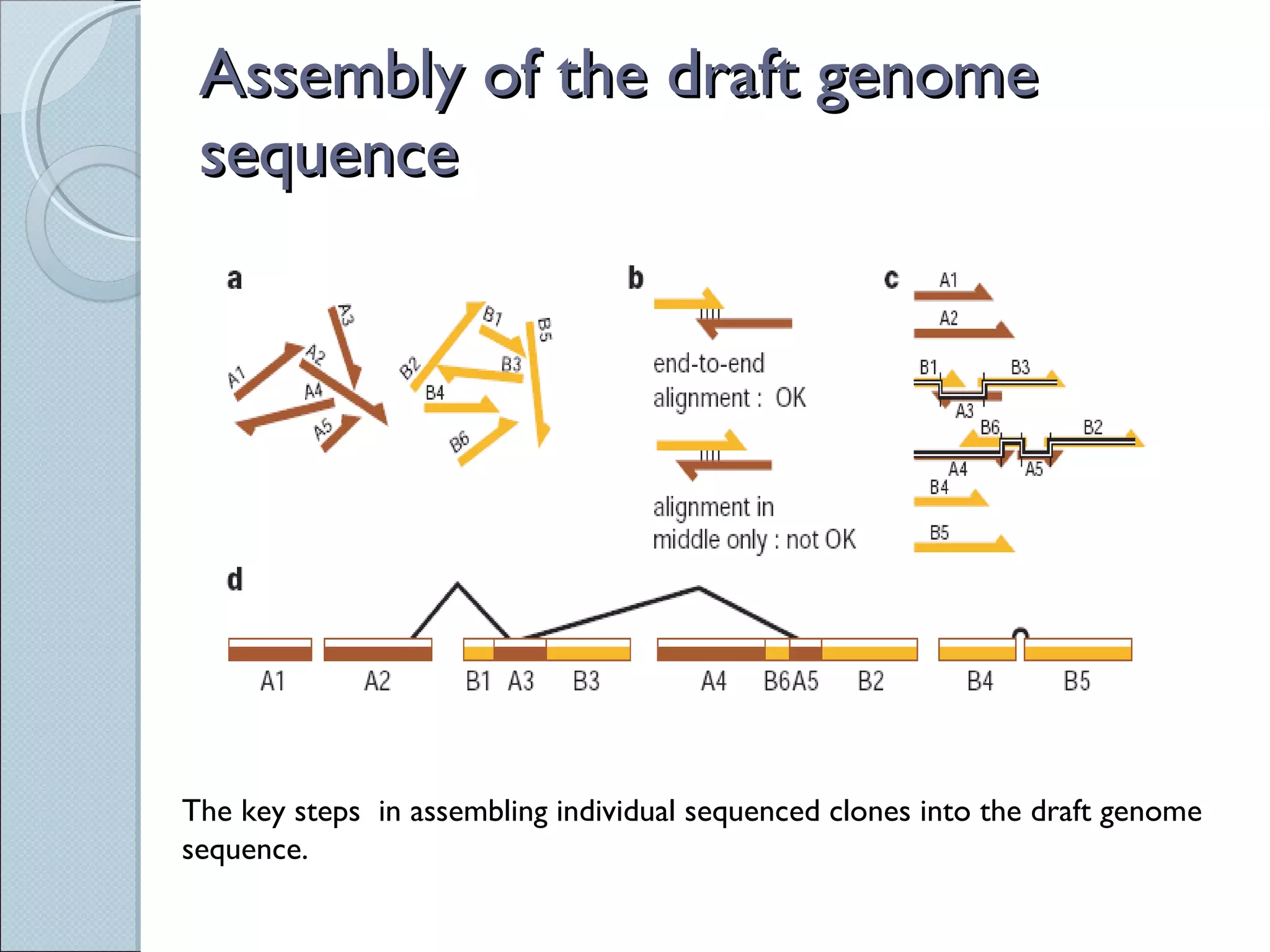
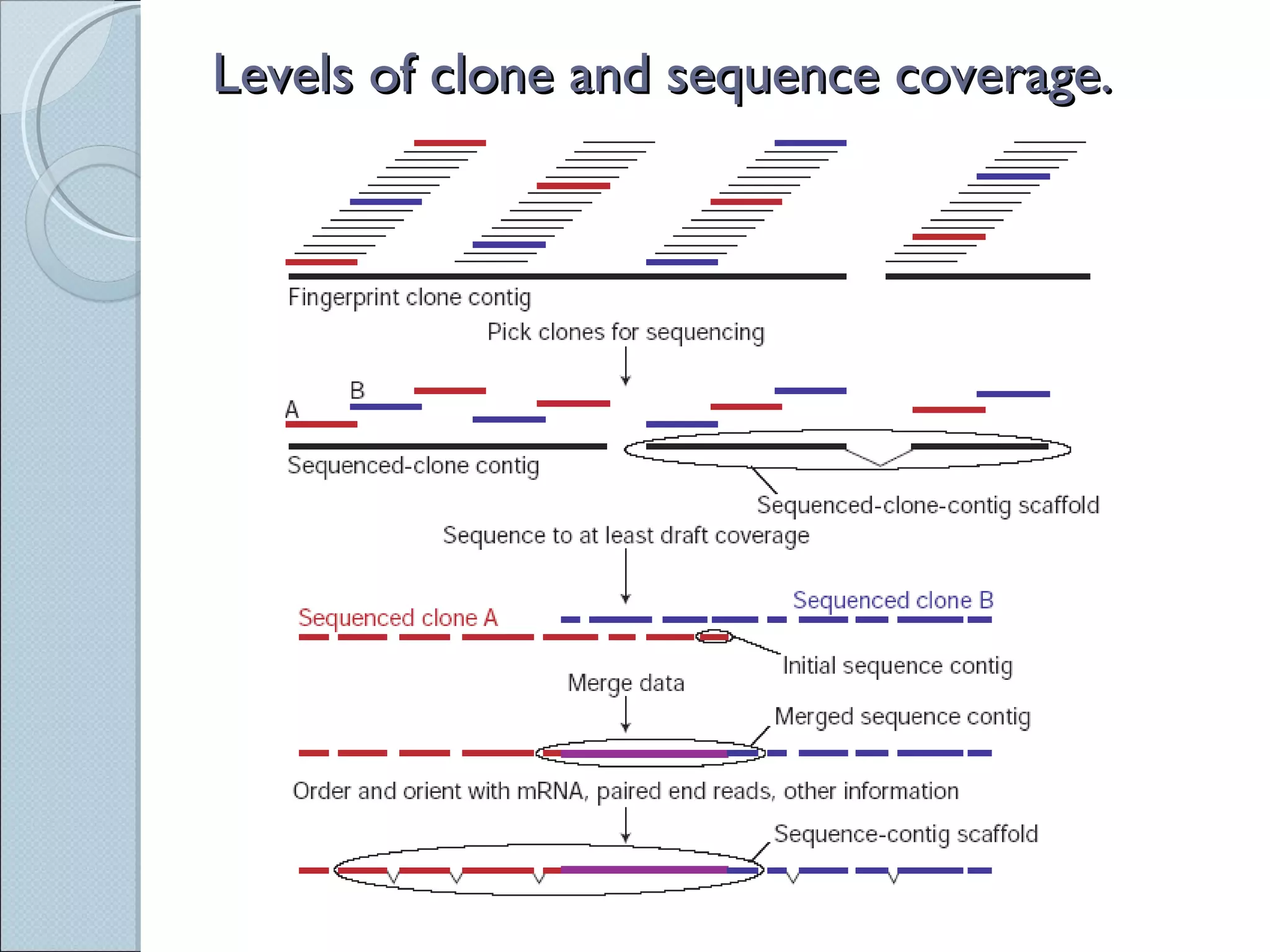
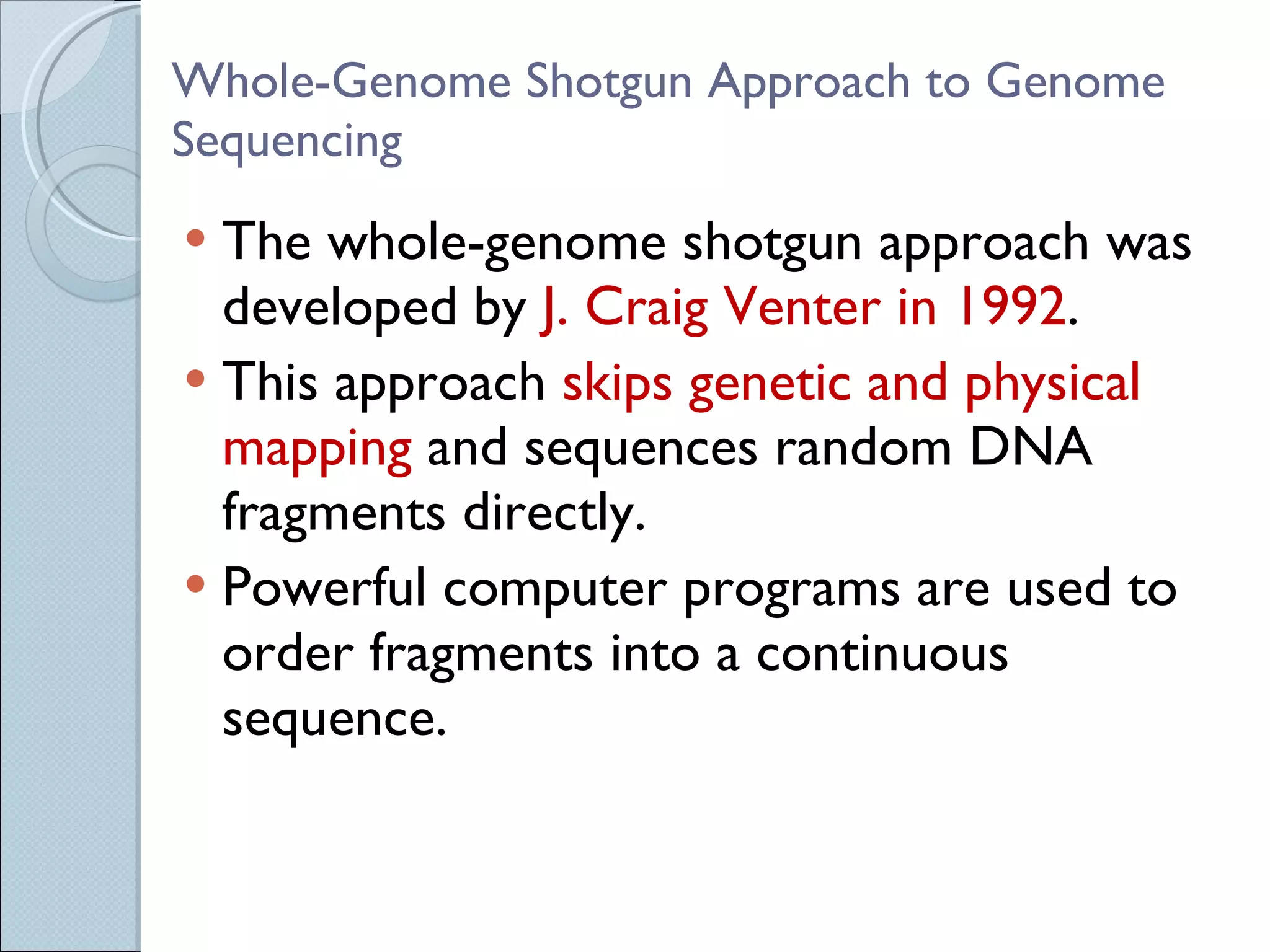
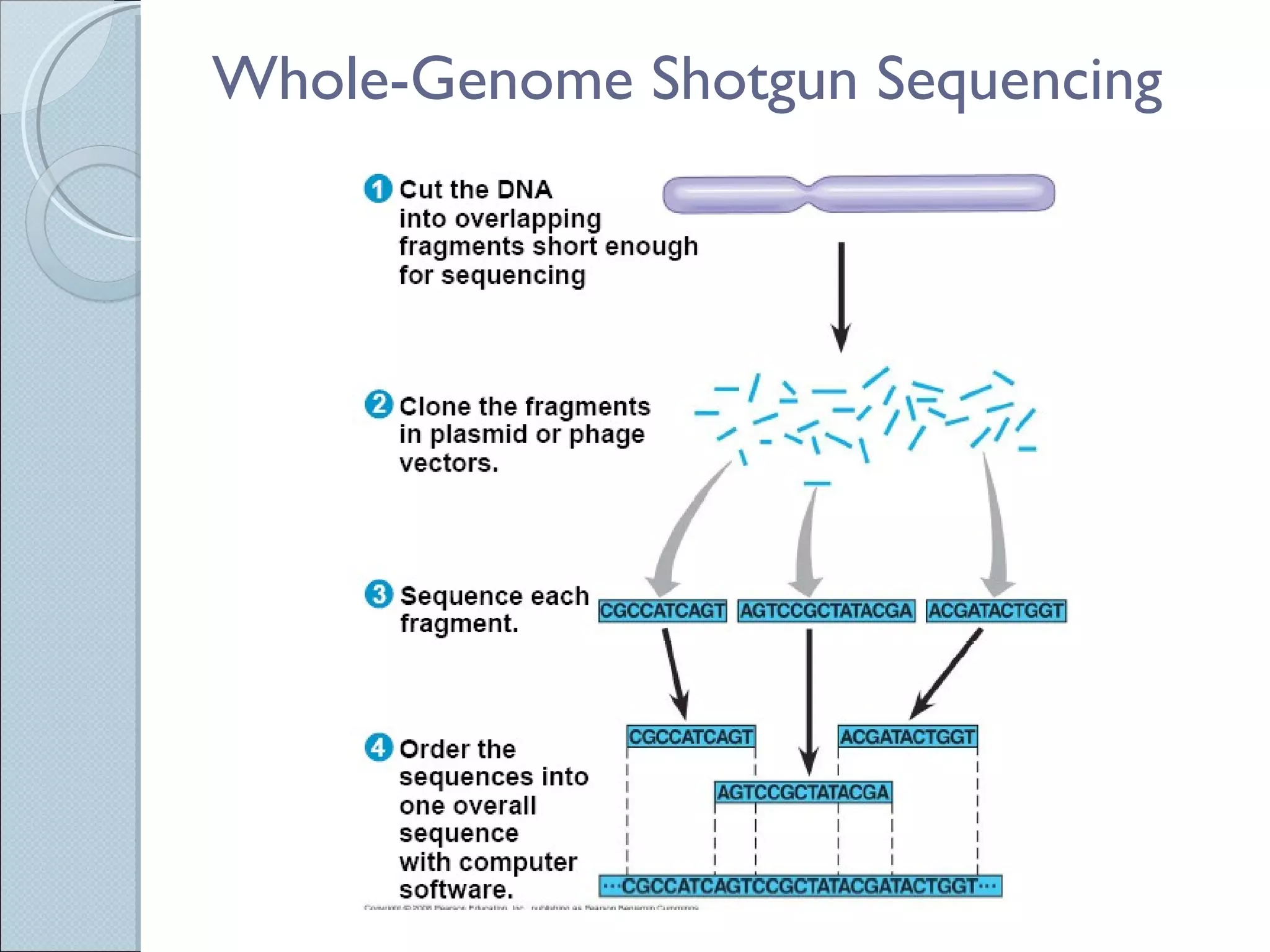
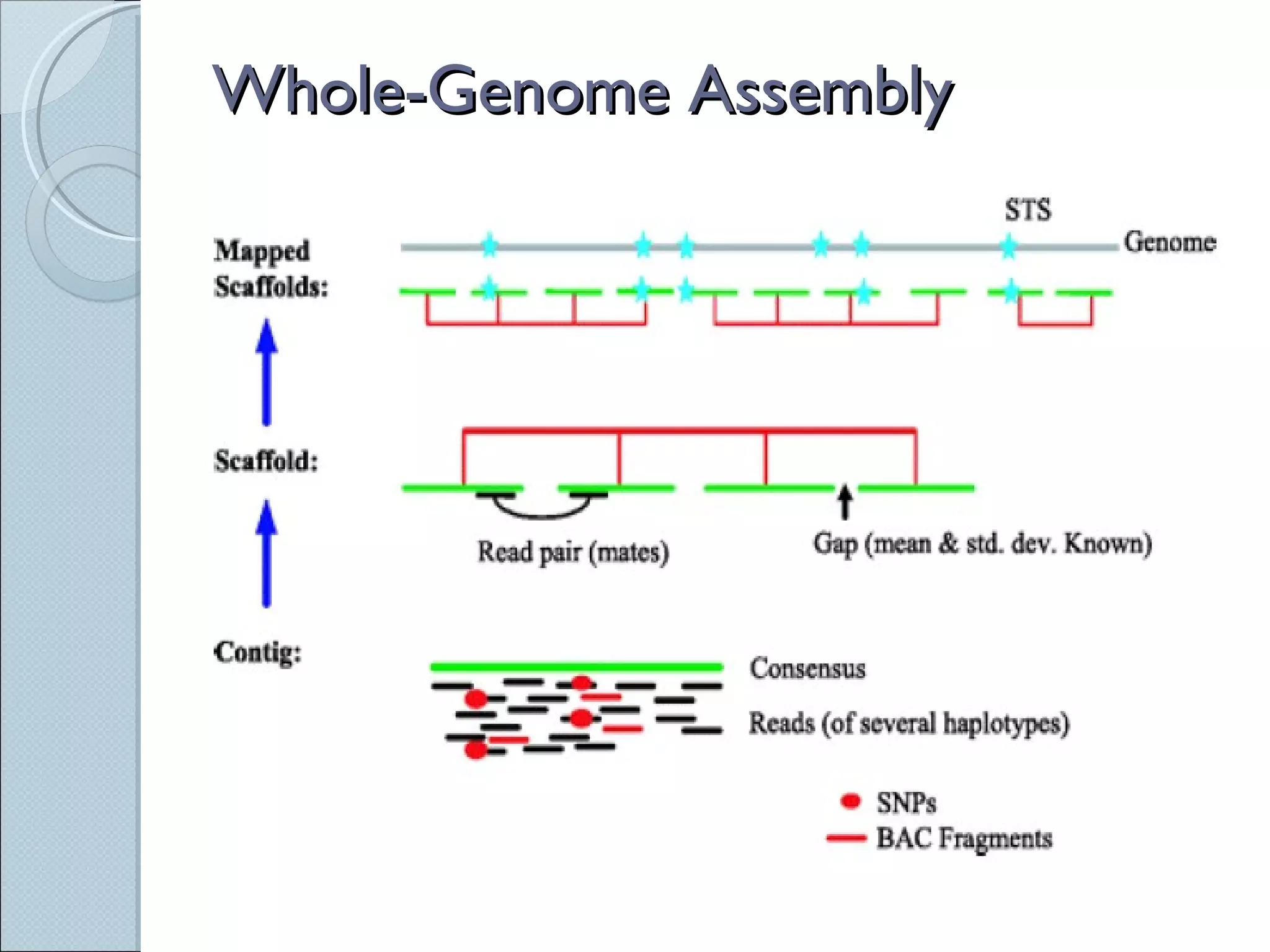
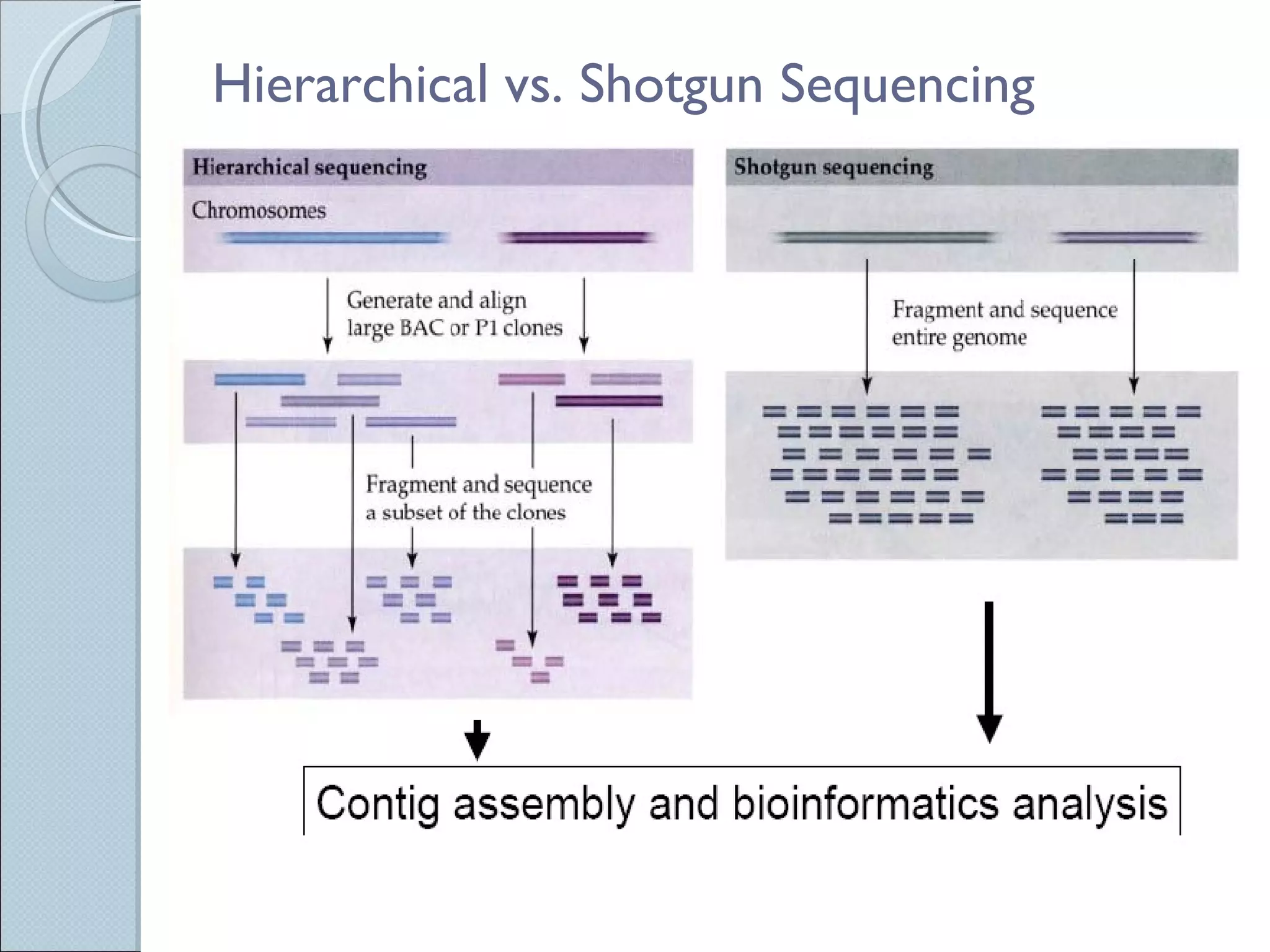
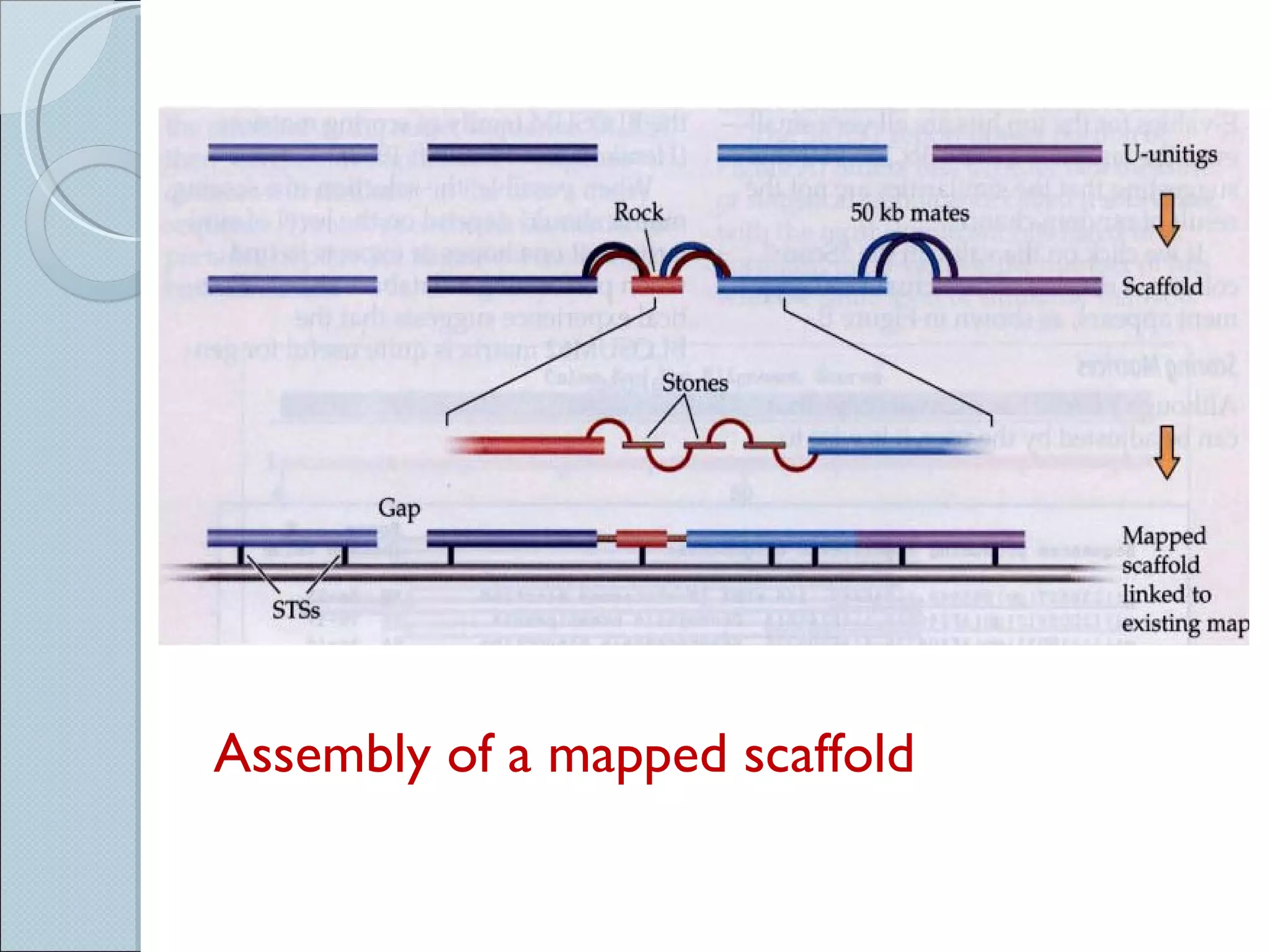
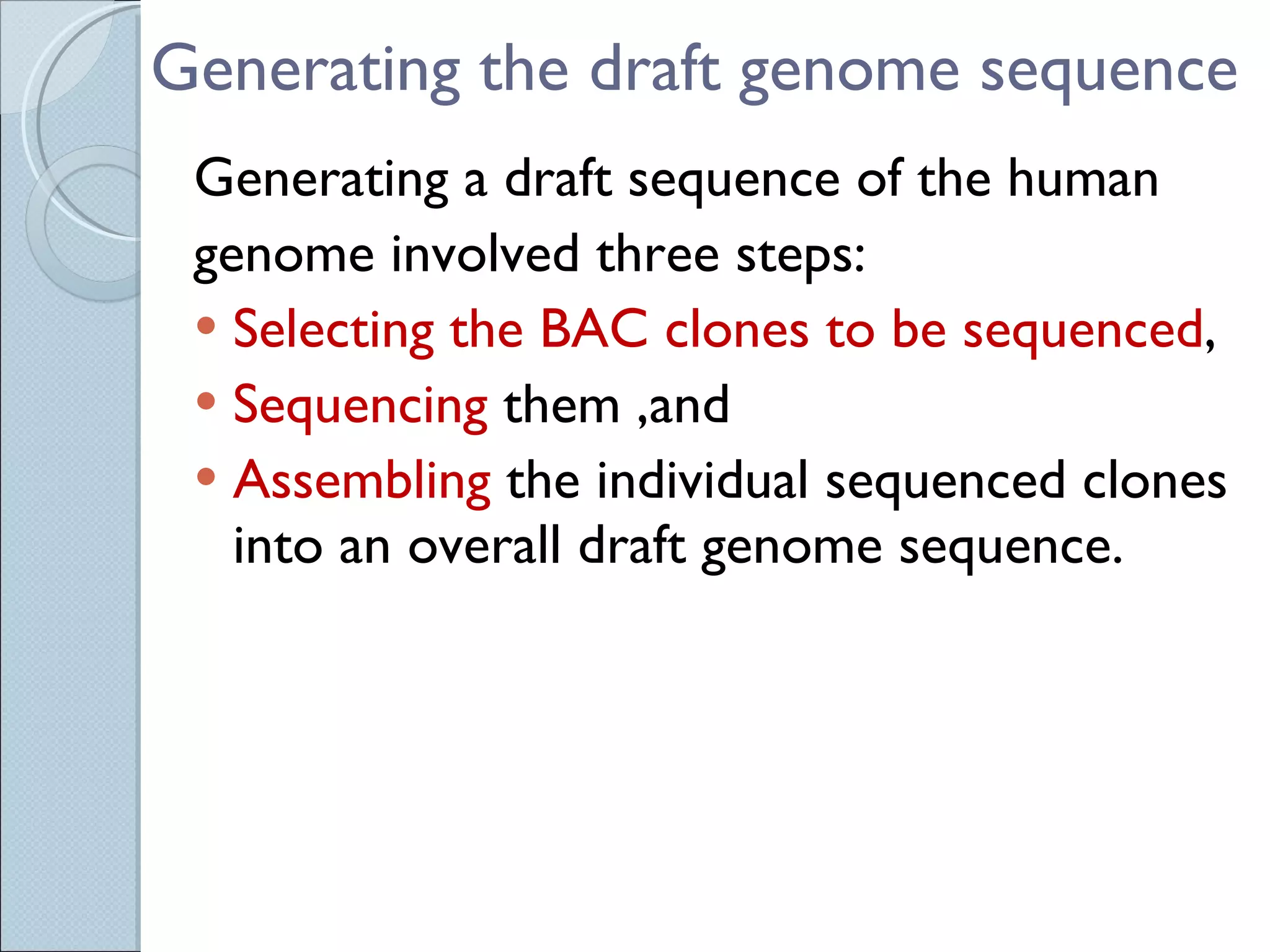
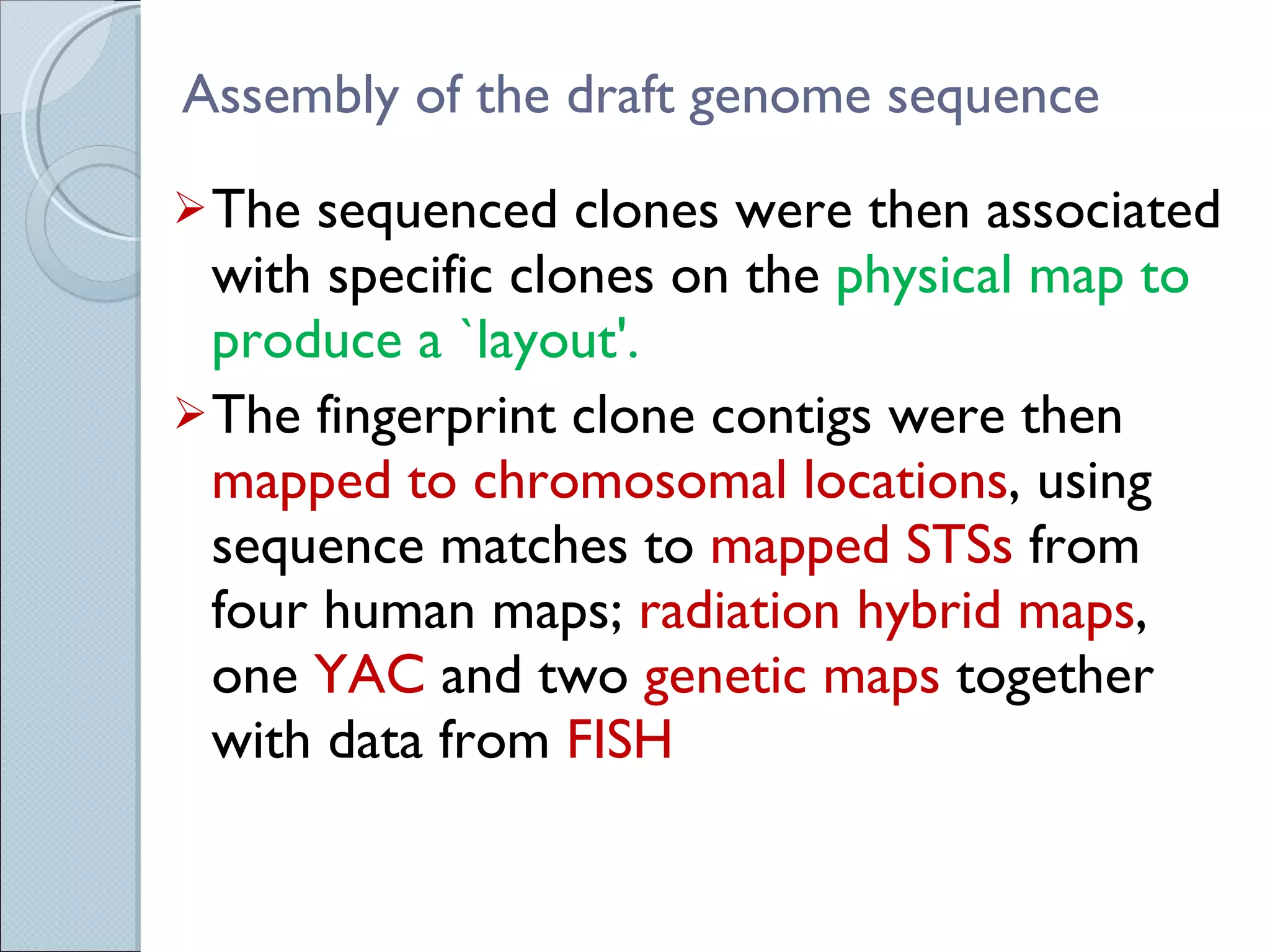
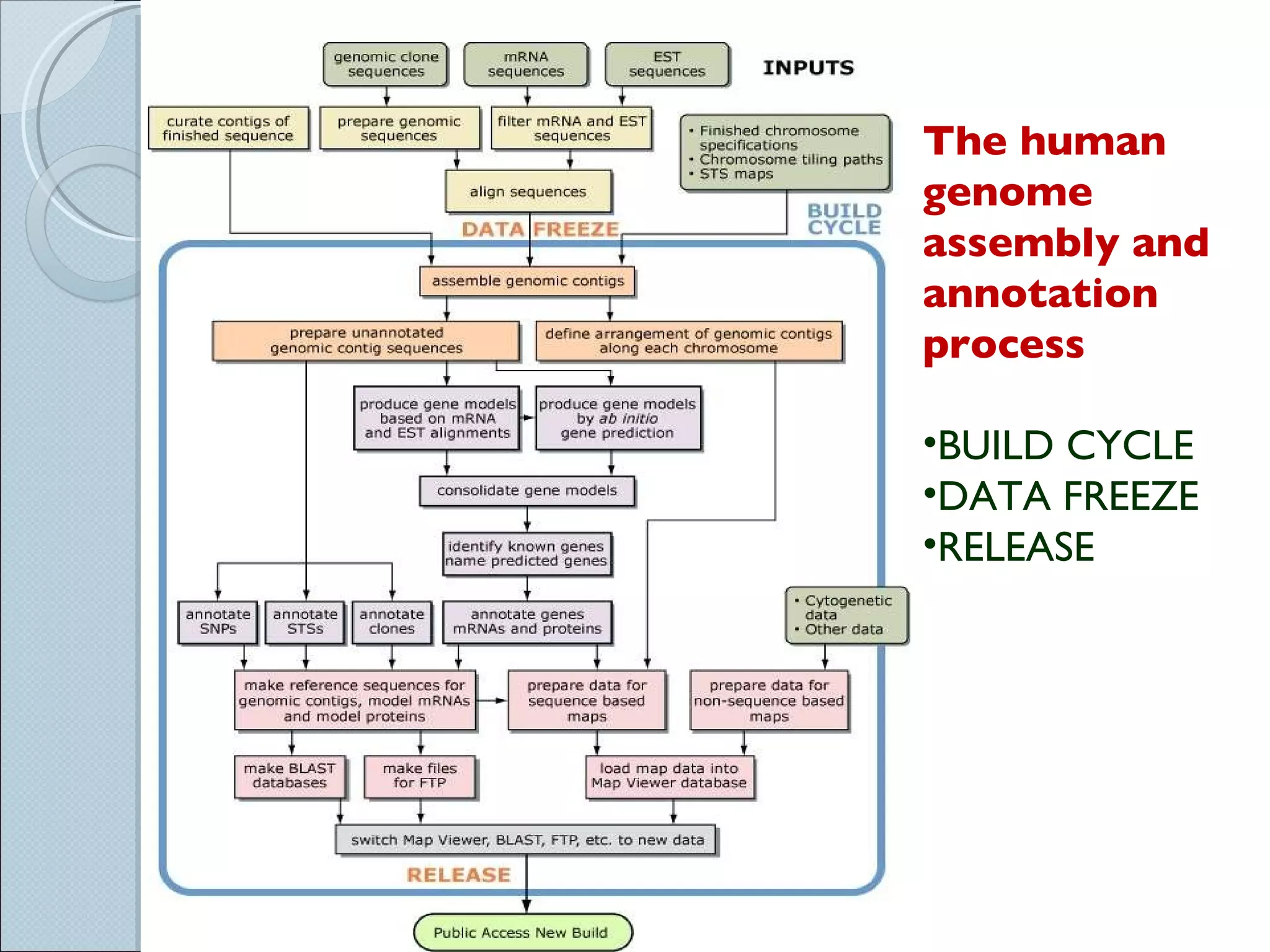
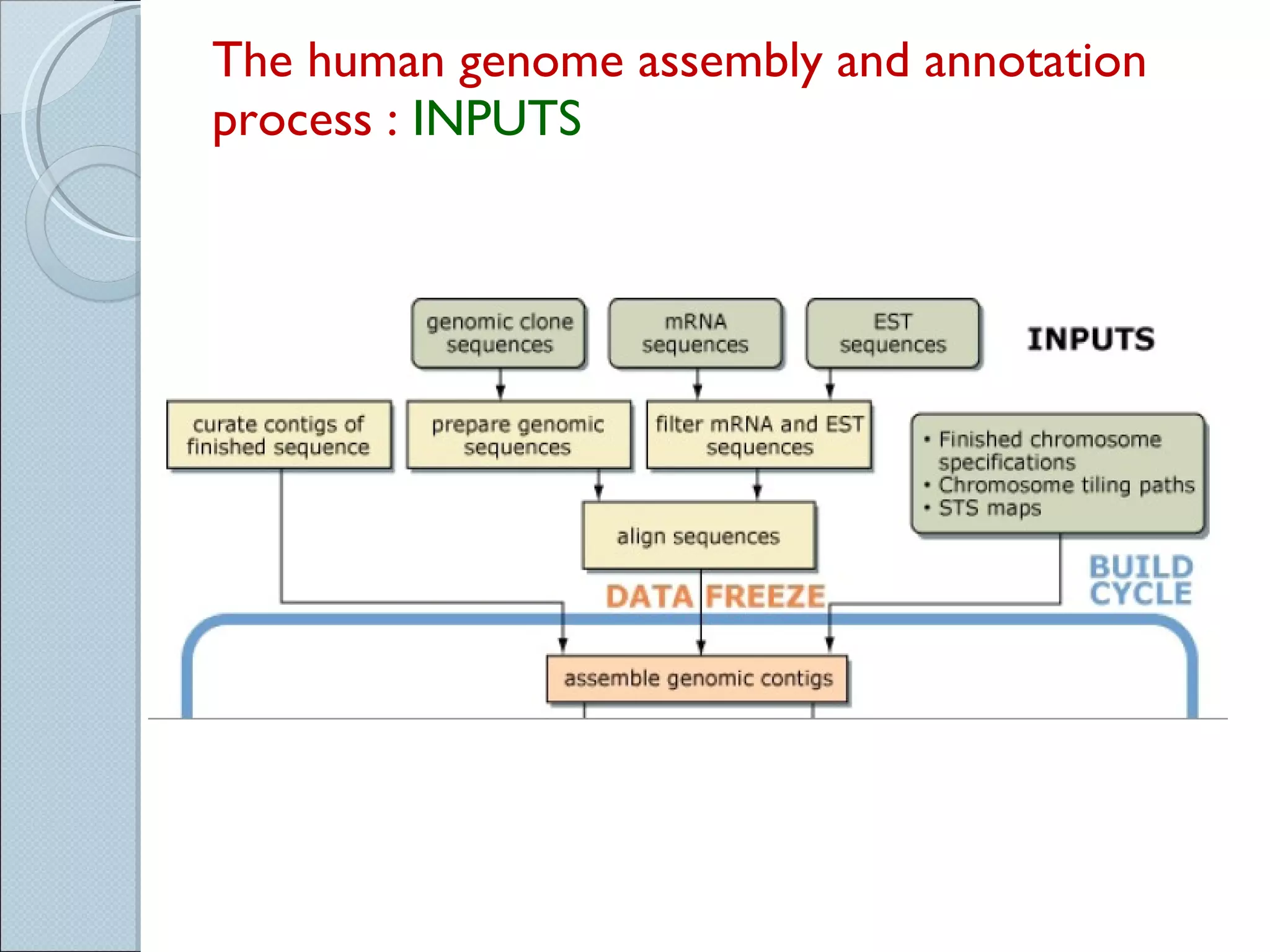
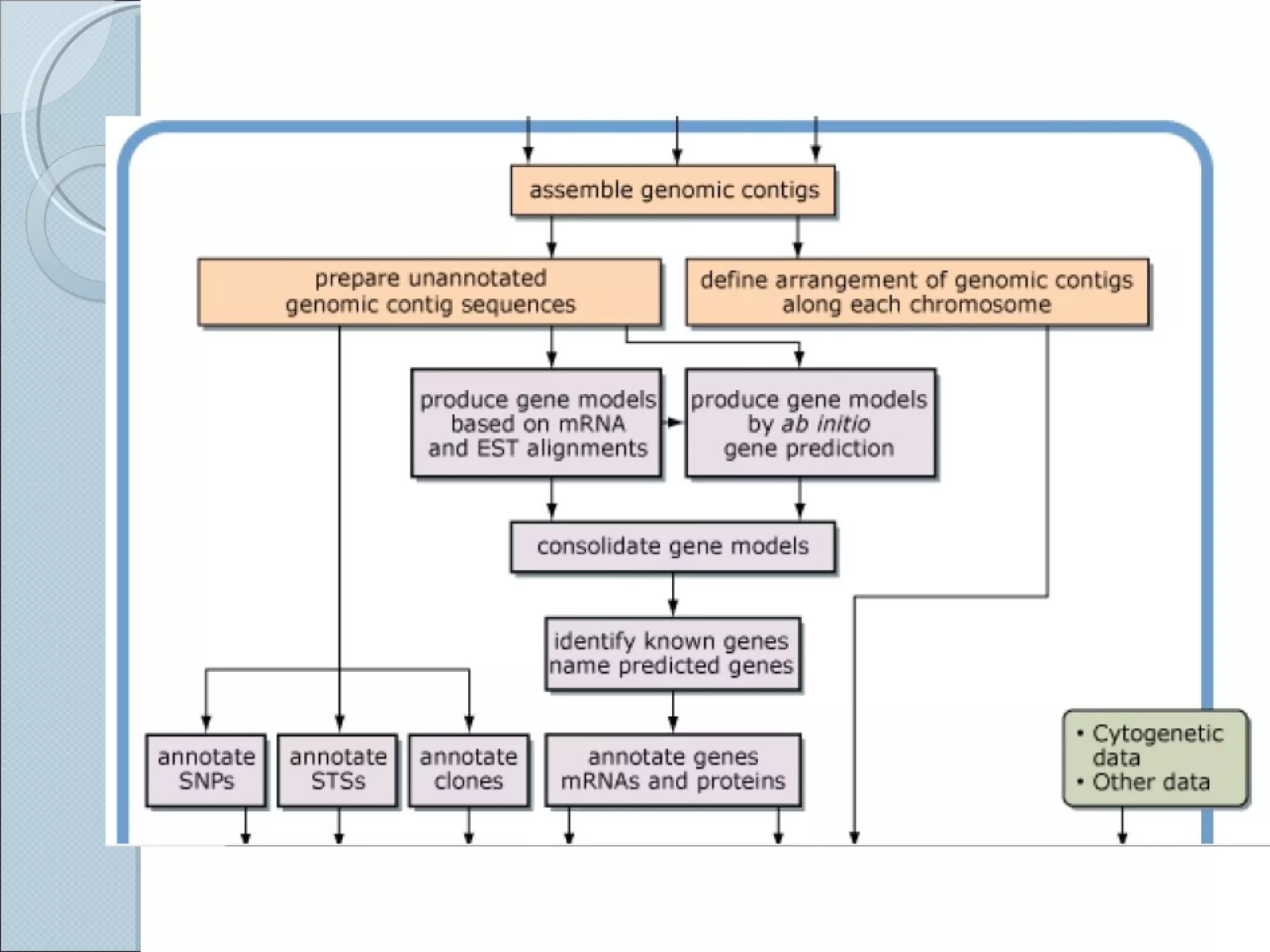
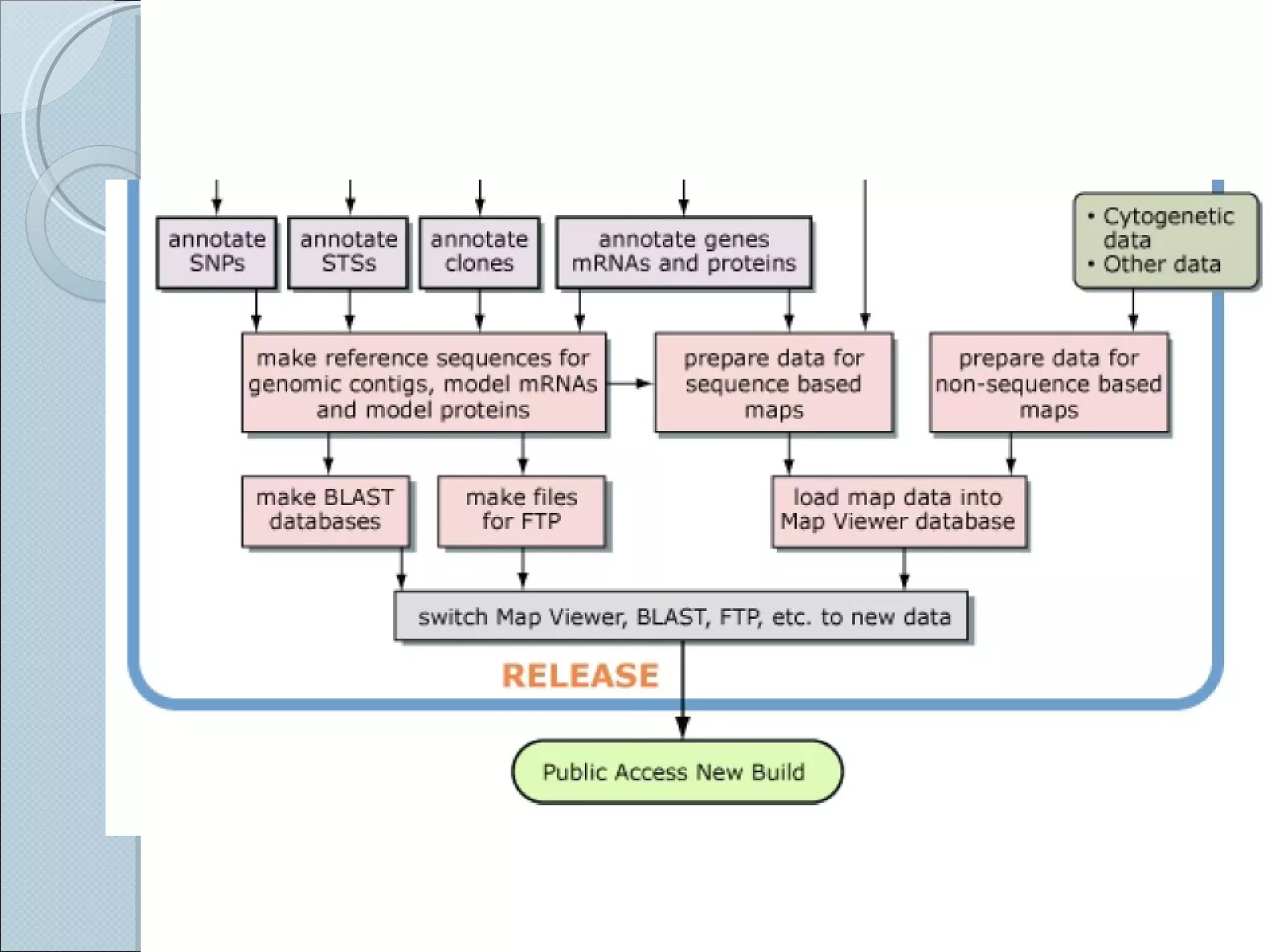
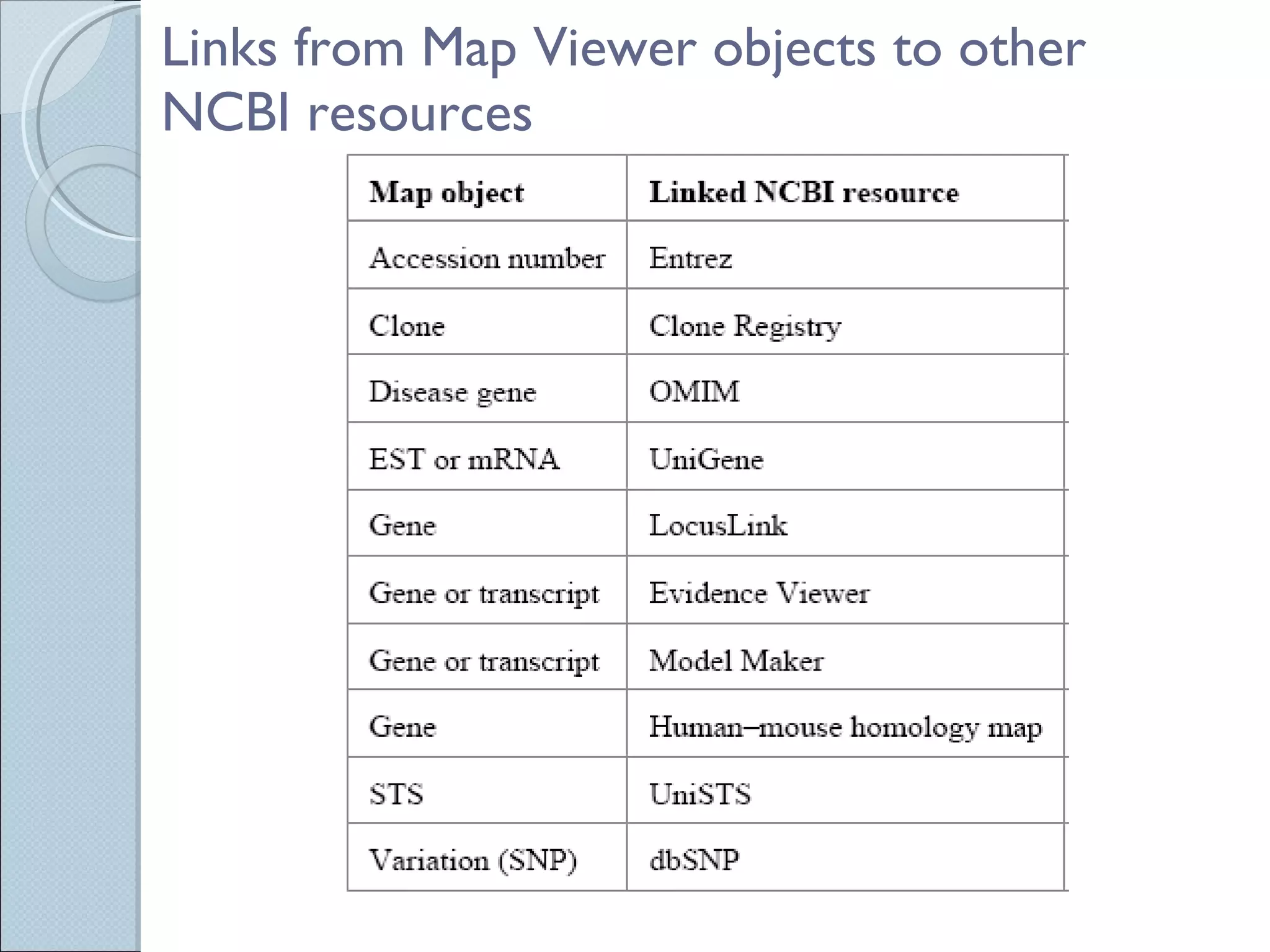
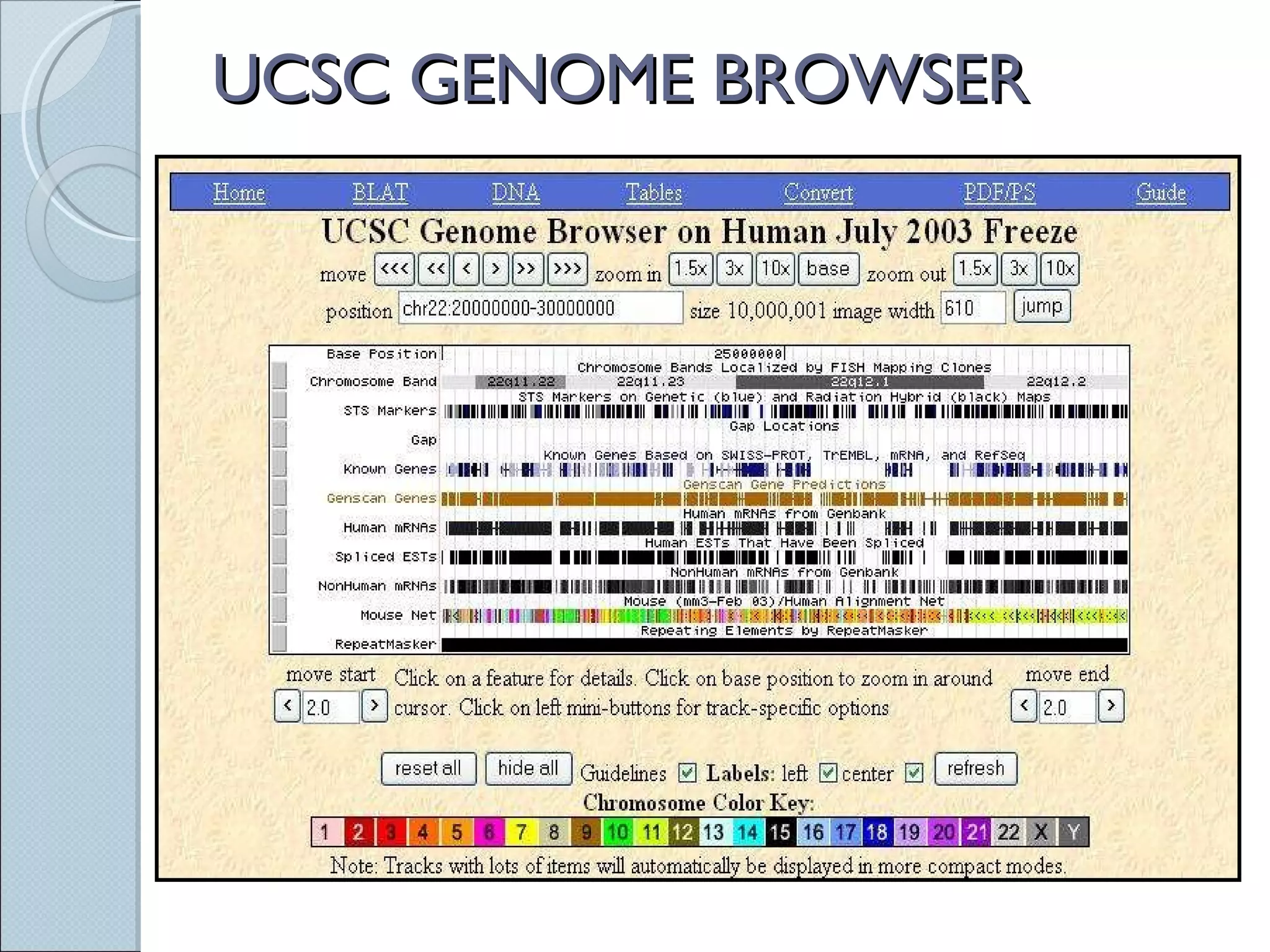
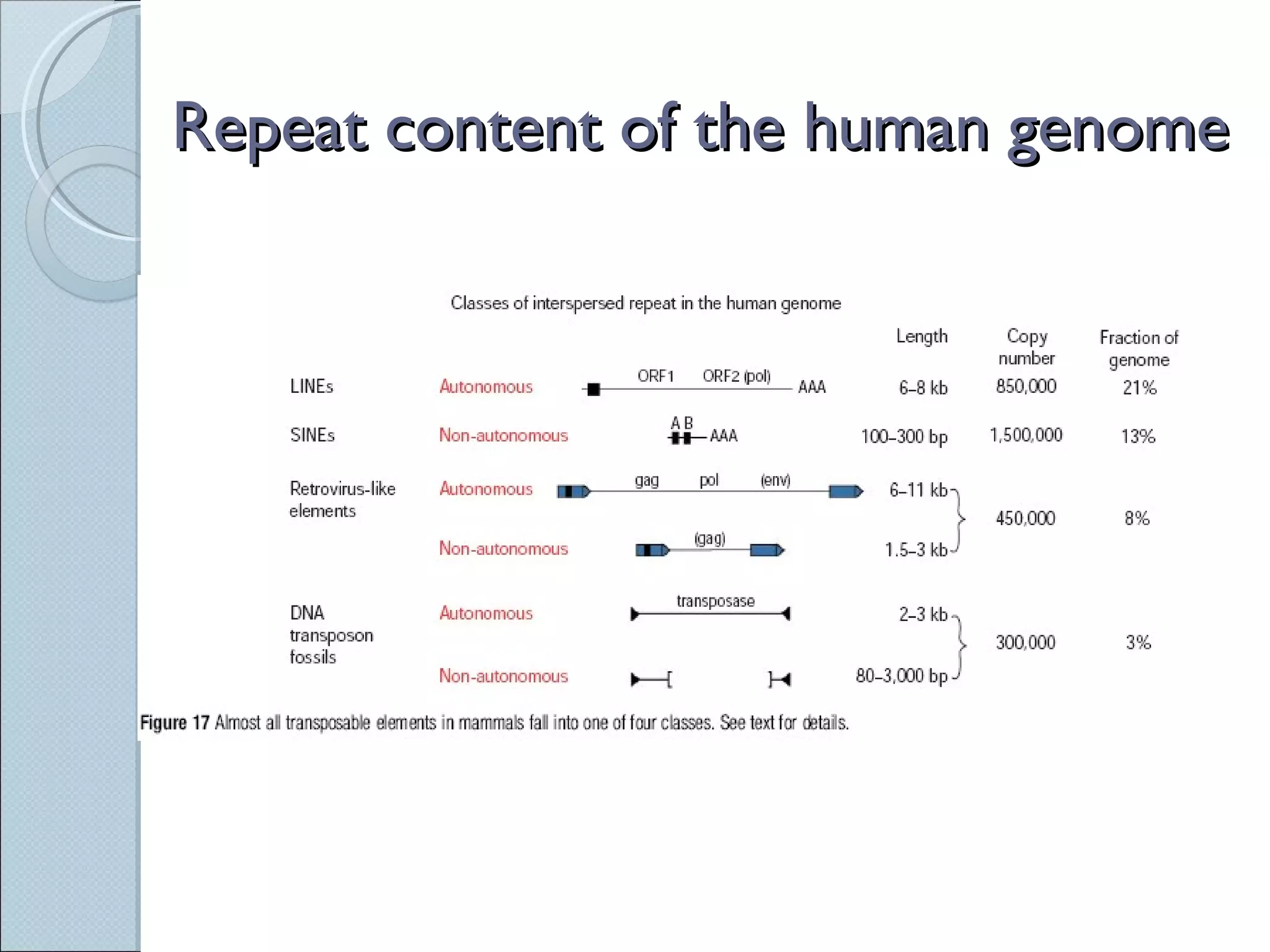
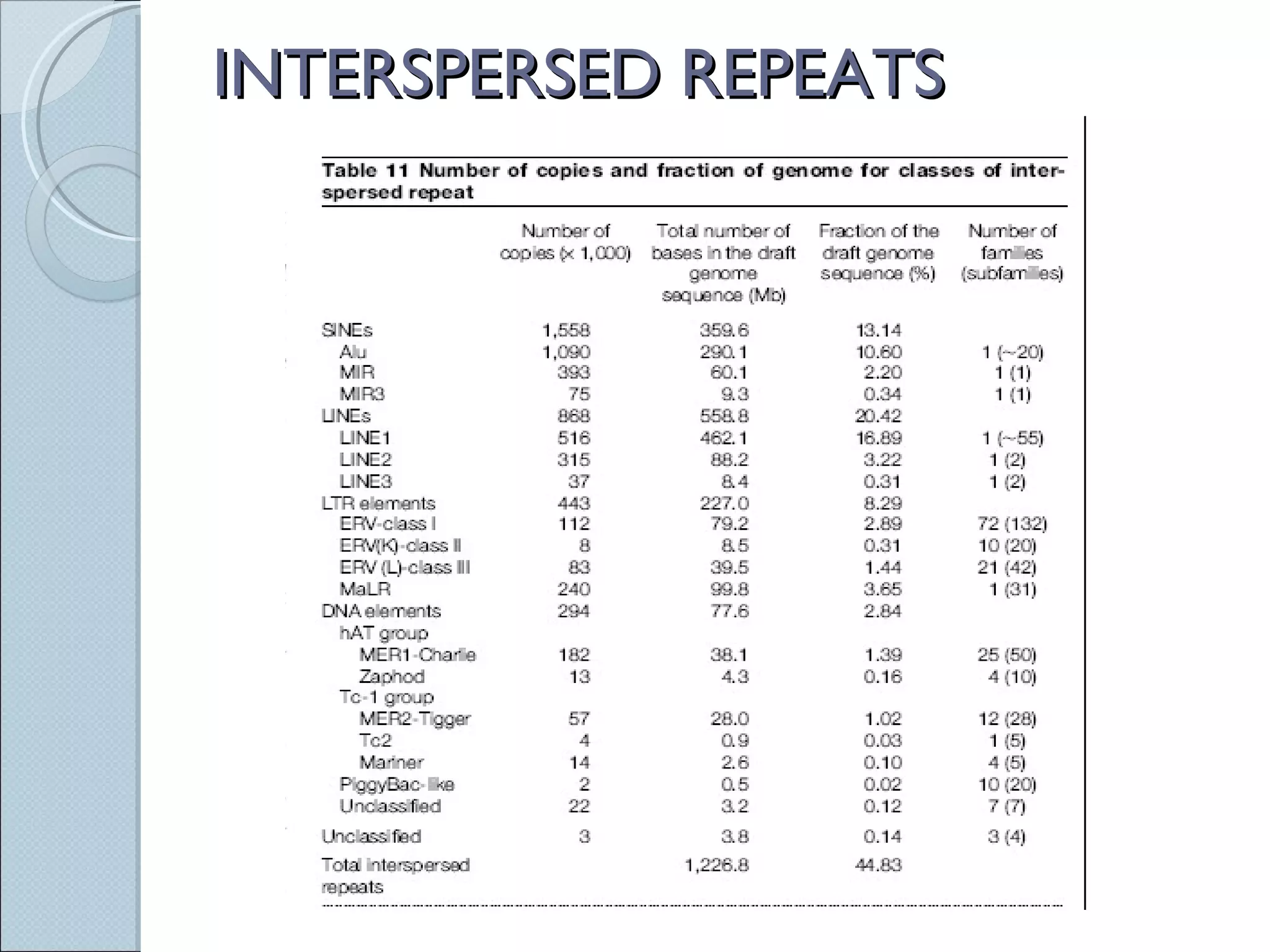
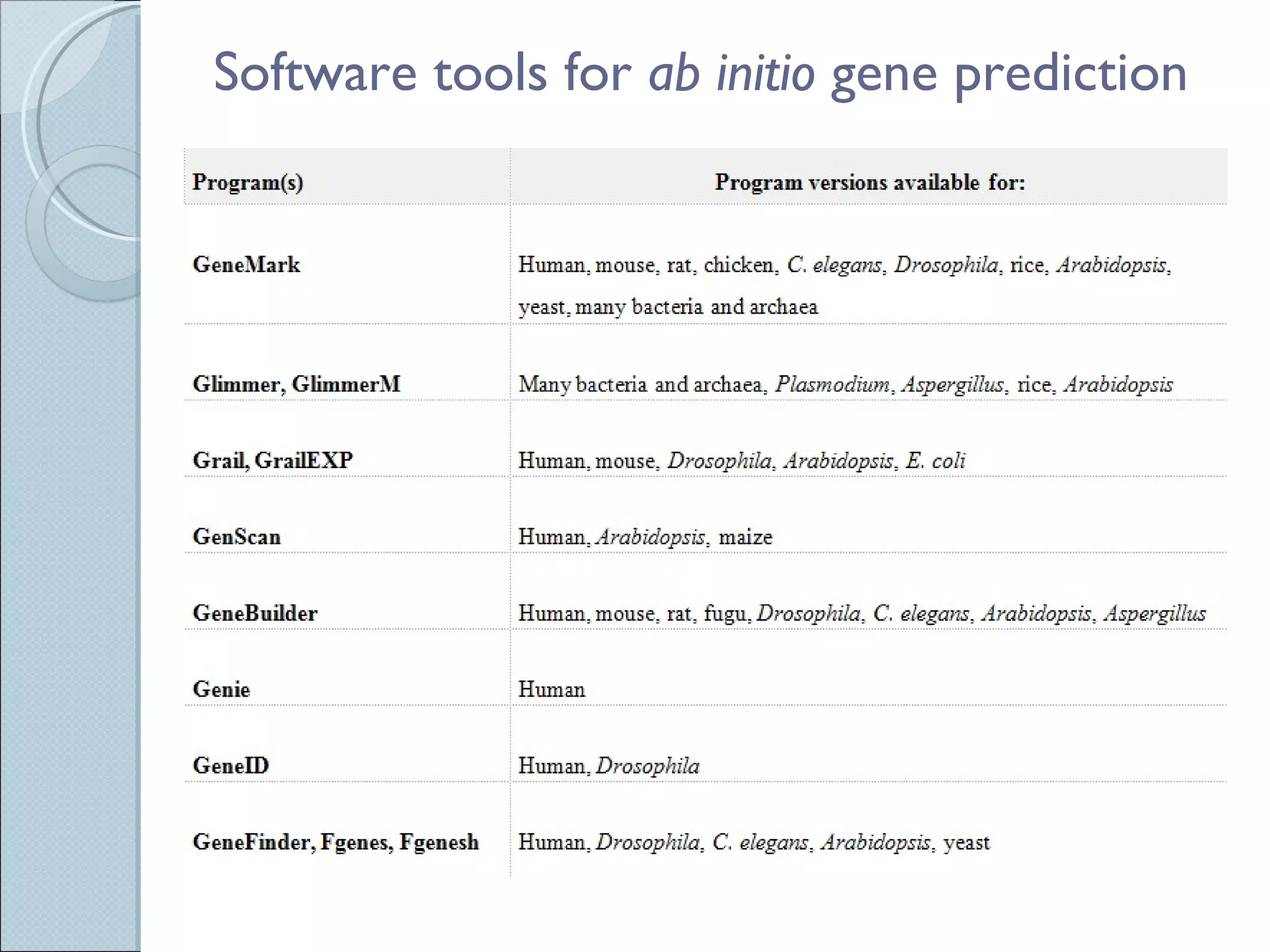
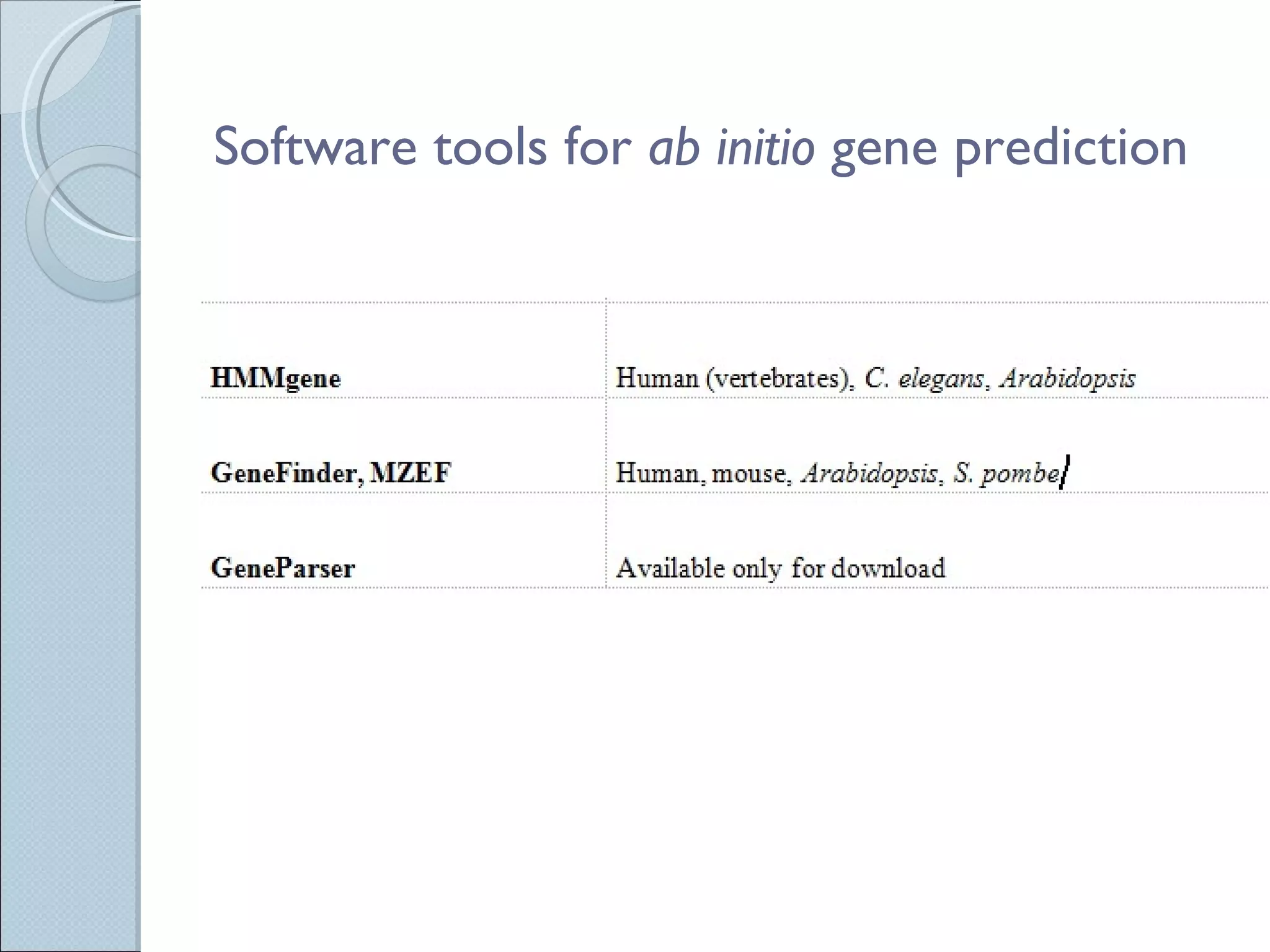
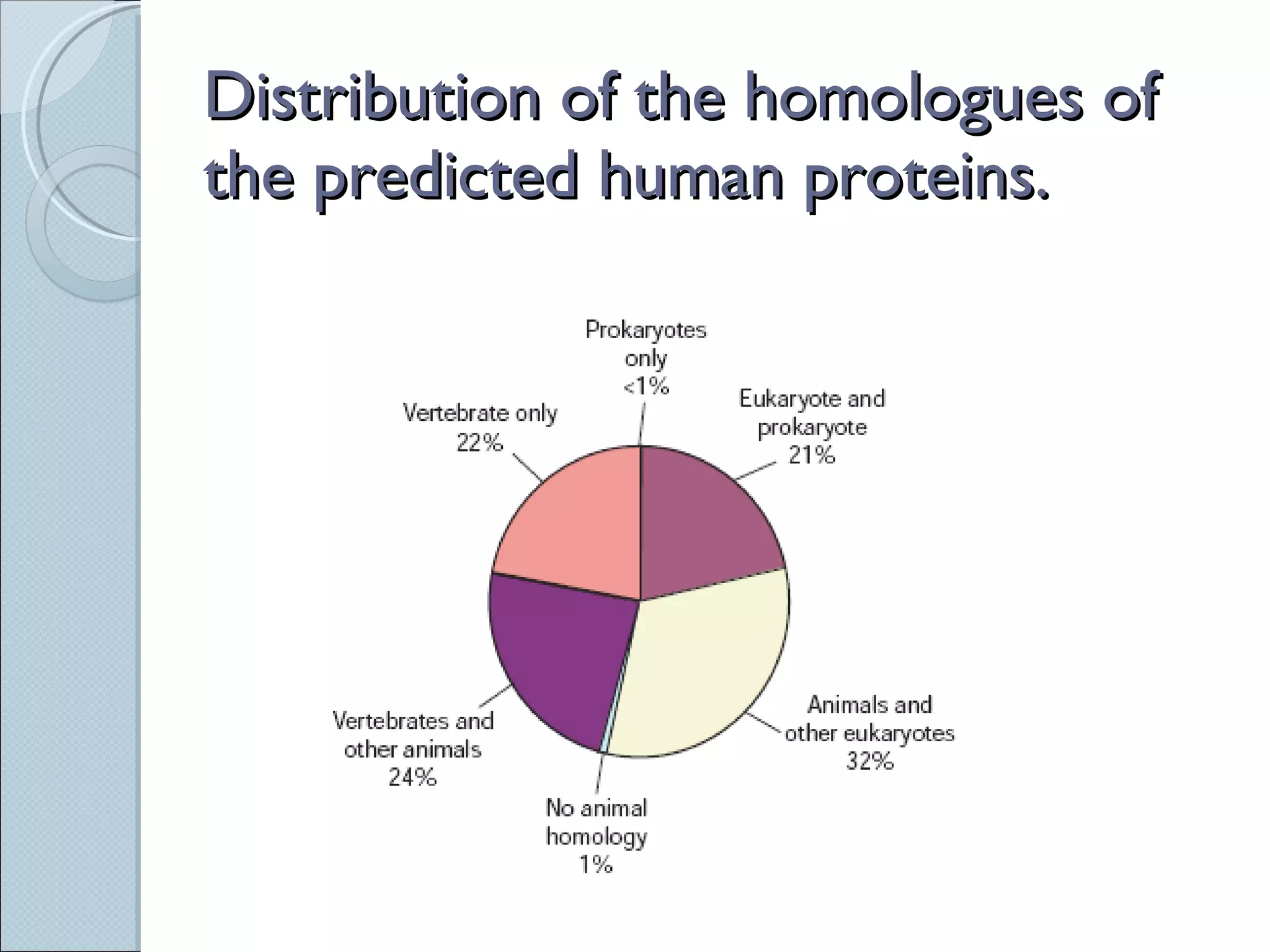
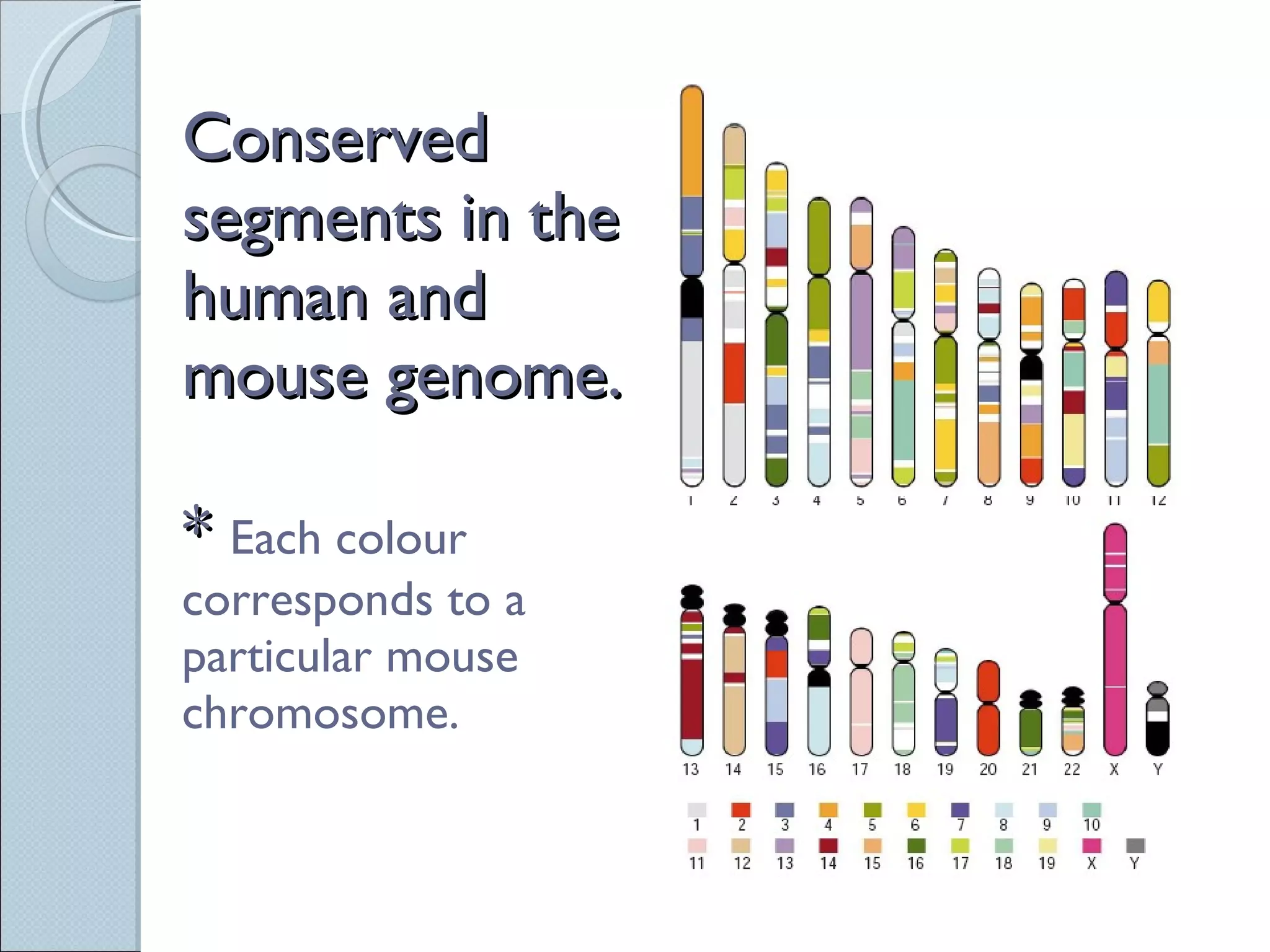
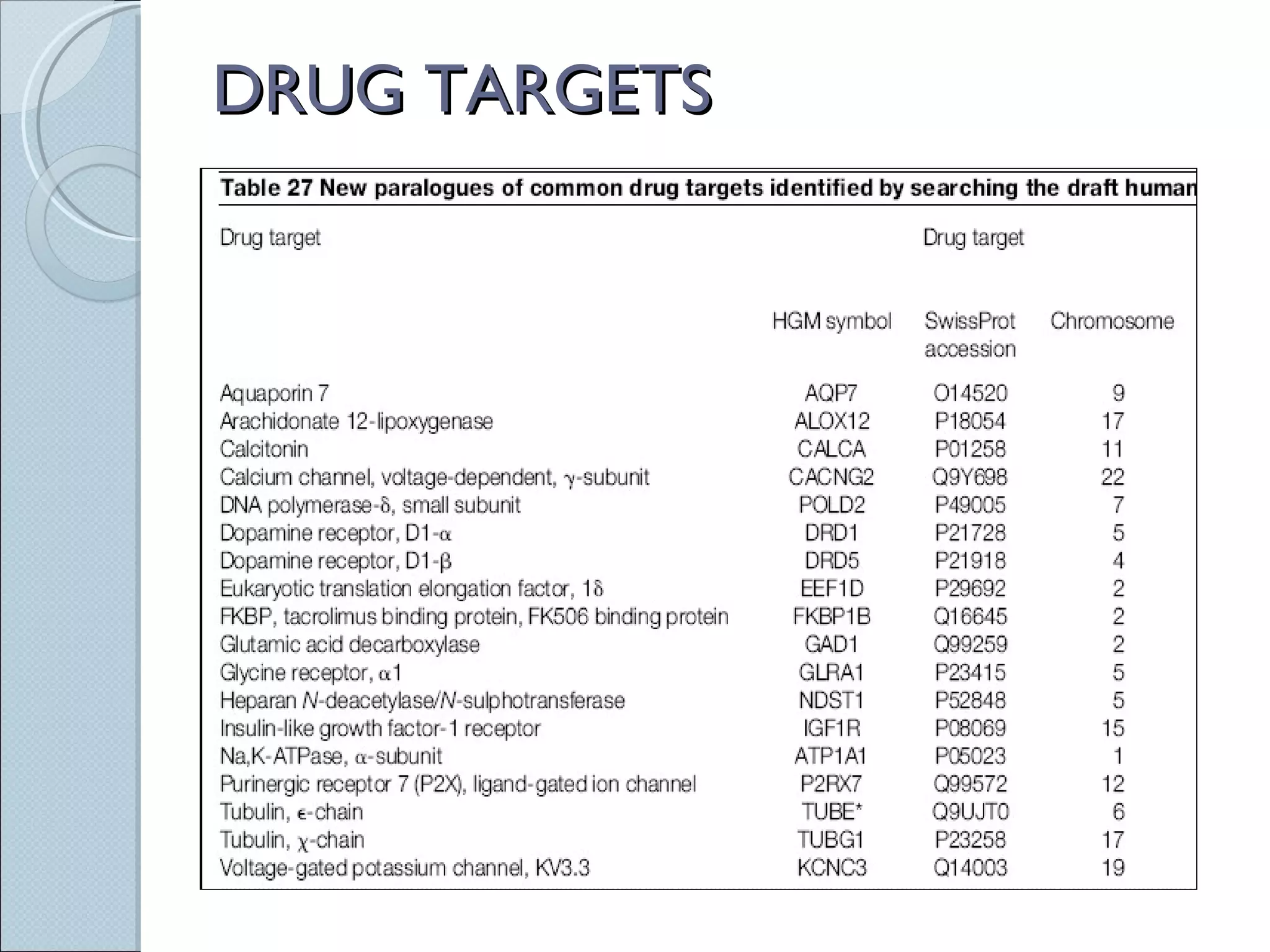
The document summarizes the Human Genome Project (HGP), which had the goals of identifying all human genes, determining the sequences of DNA base pairs that make up human DNA, storing this information in databases, and addressing ethical issues. Key milestones included completing a working draft in 2000 and fully sequencing the genome in 2003, two years ahead of schedule. The HGP and a private company collaborated to obtain the DNA sequence.