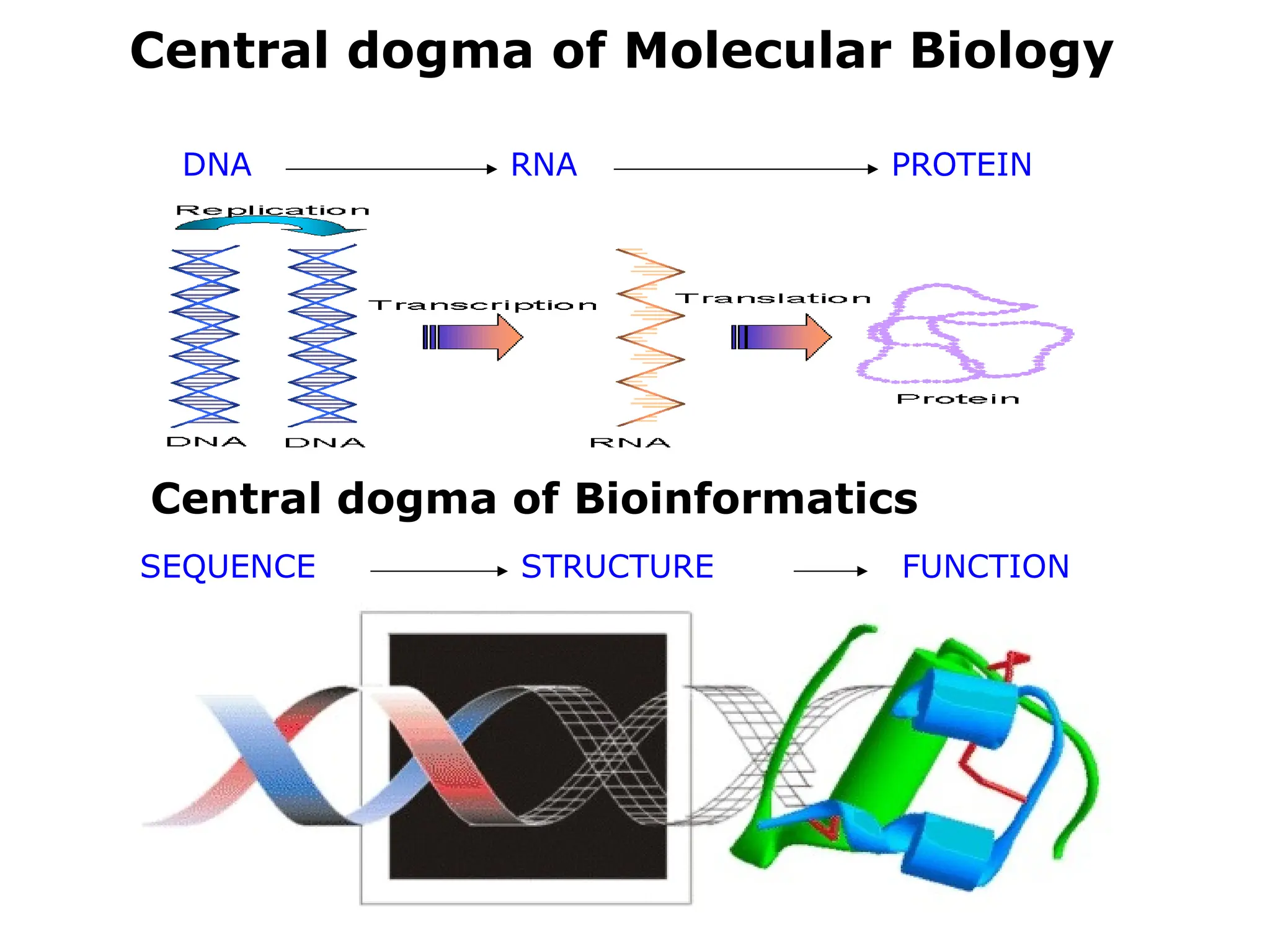
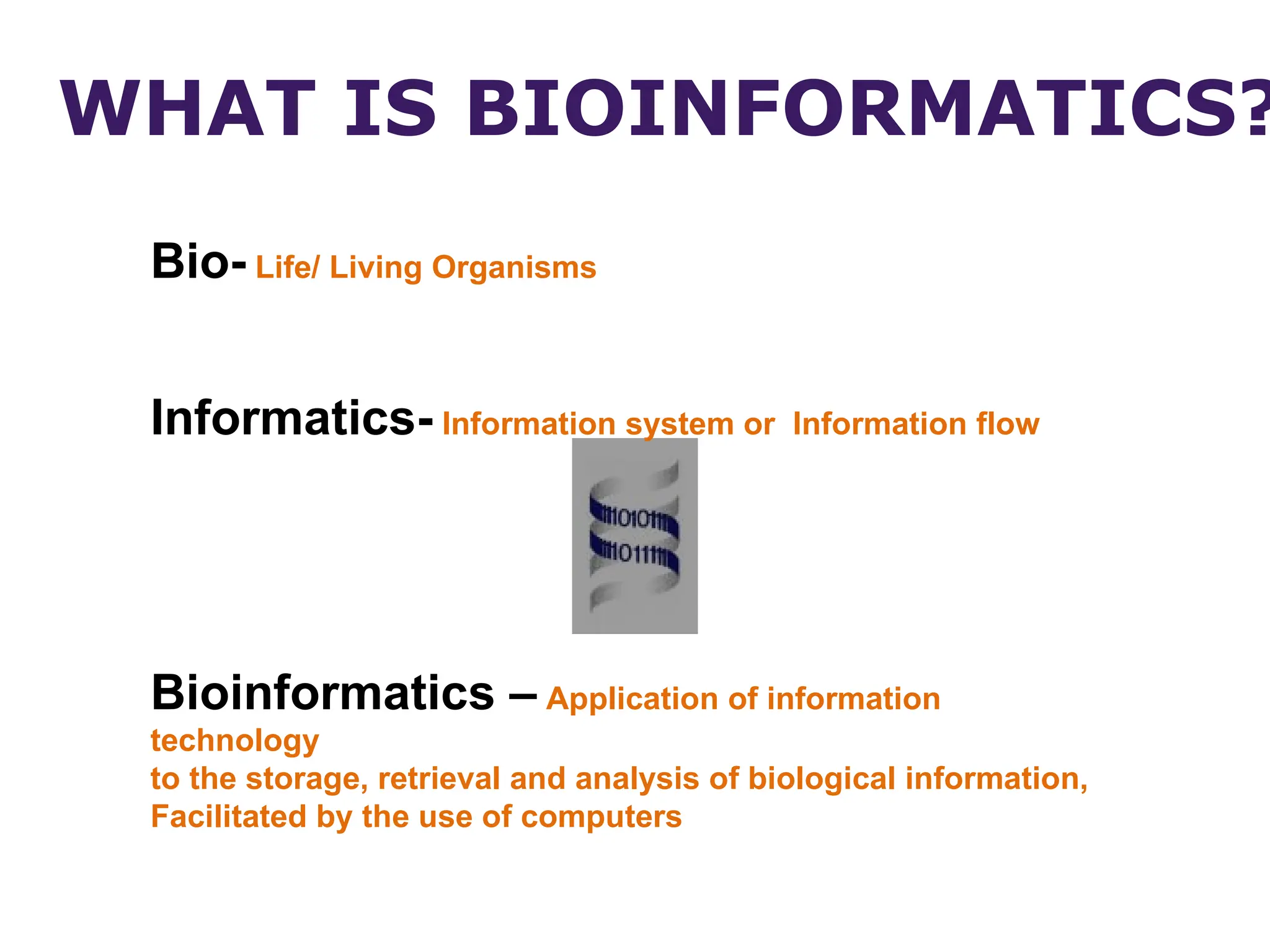
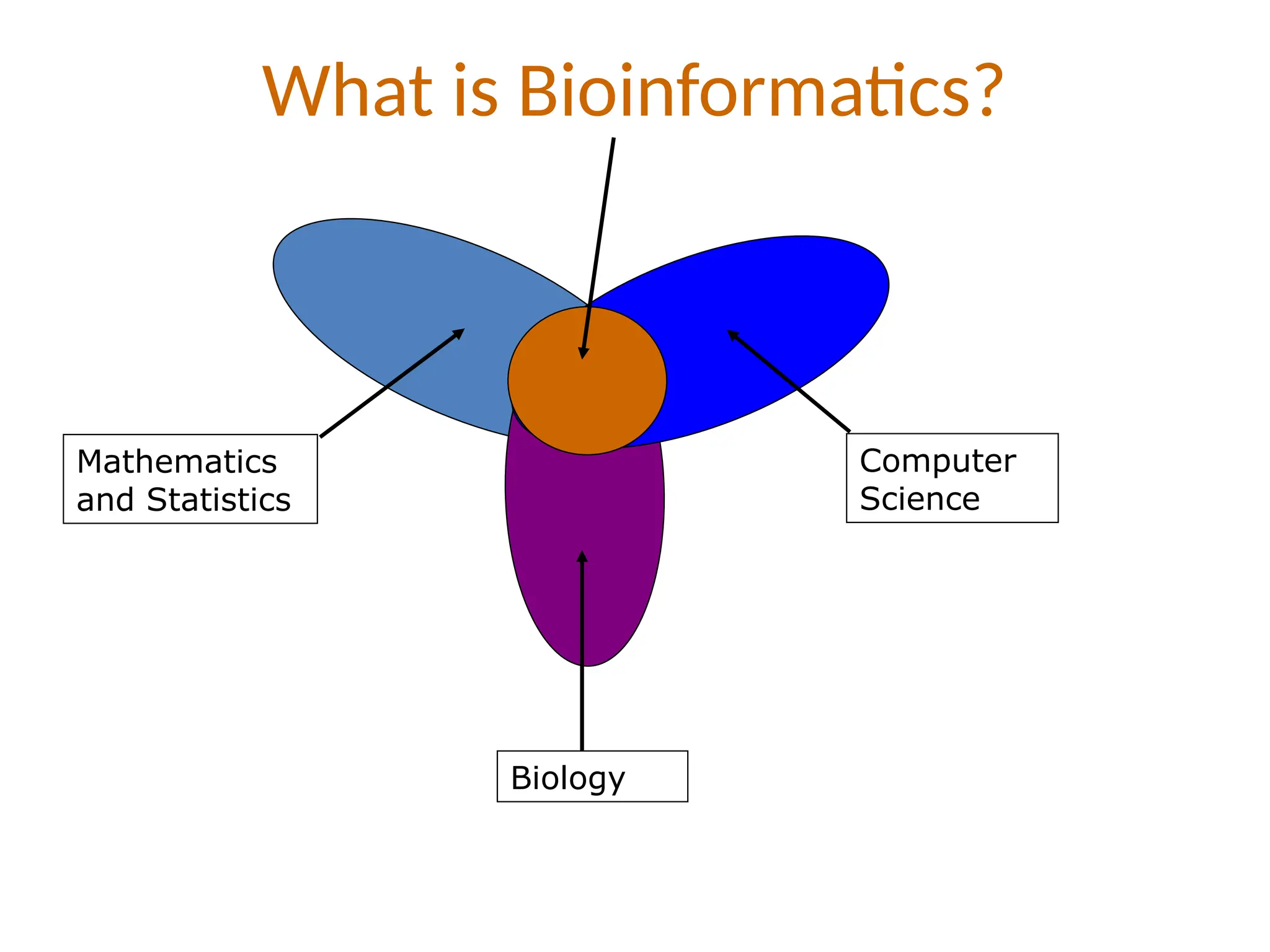
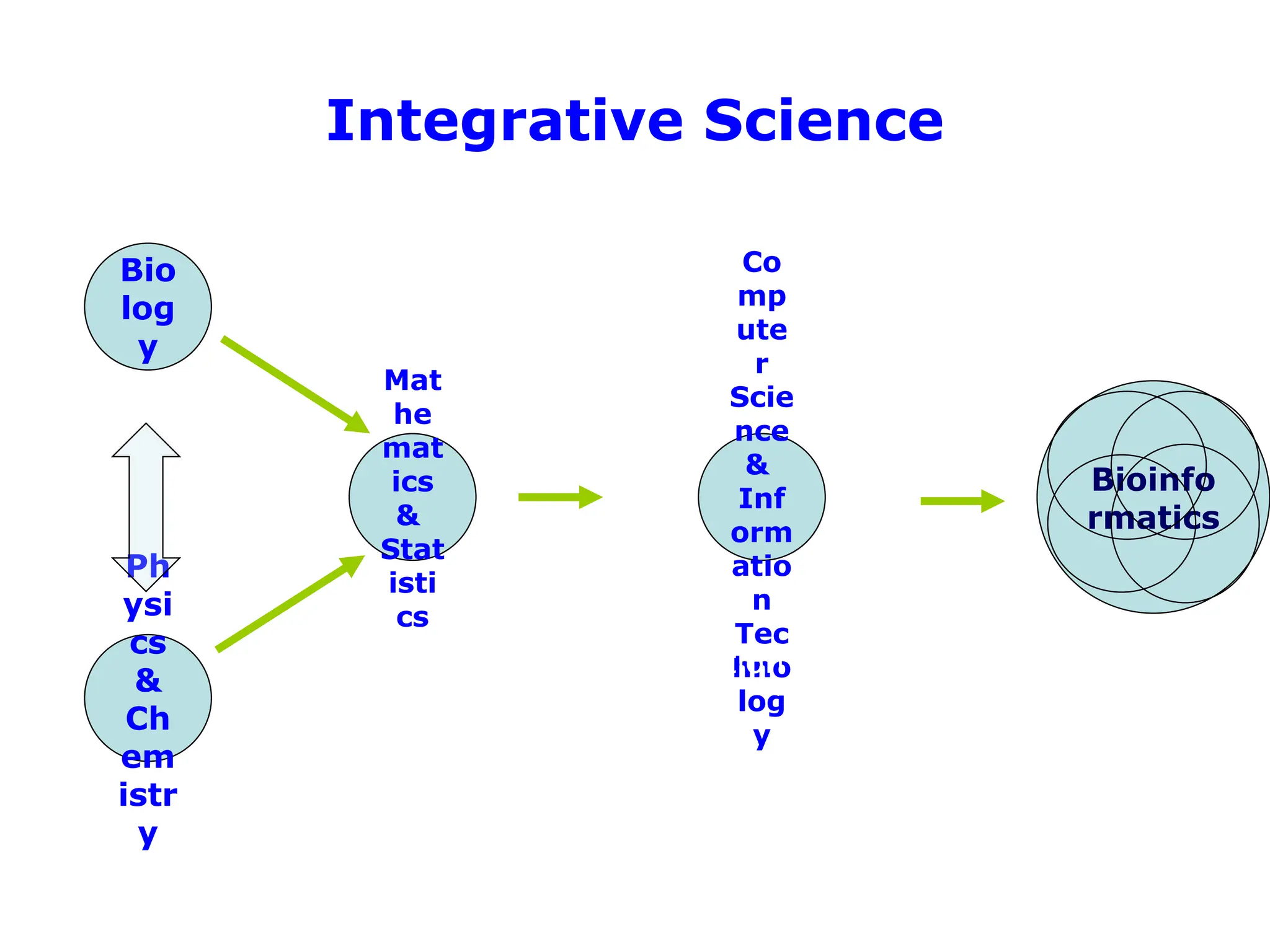
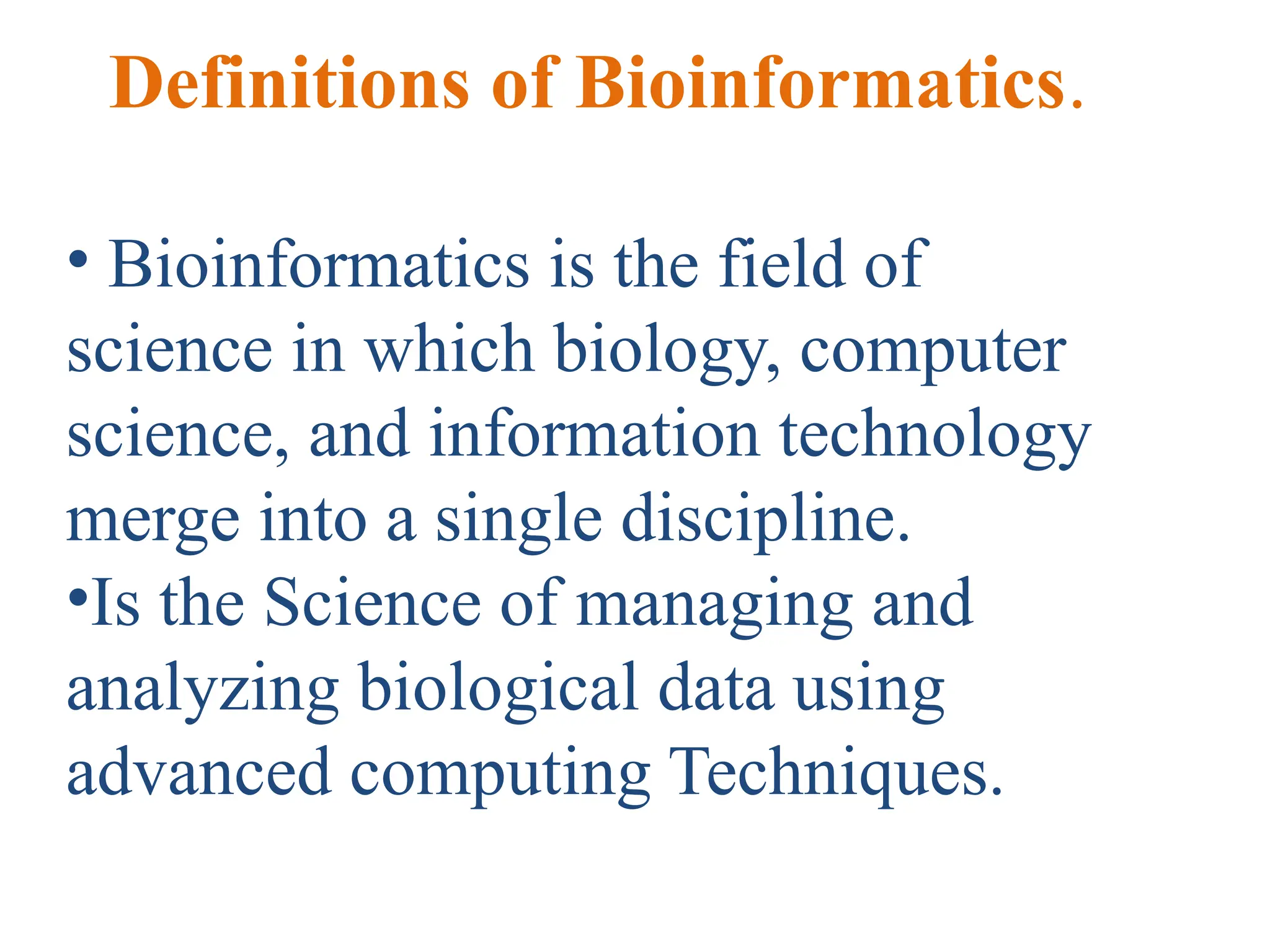
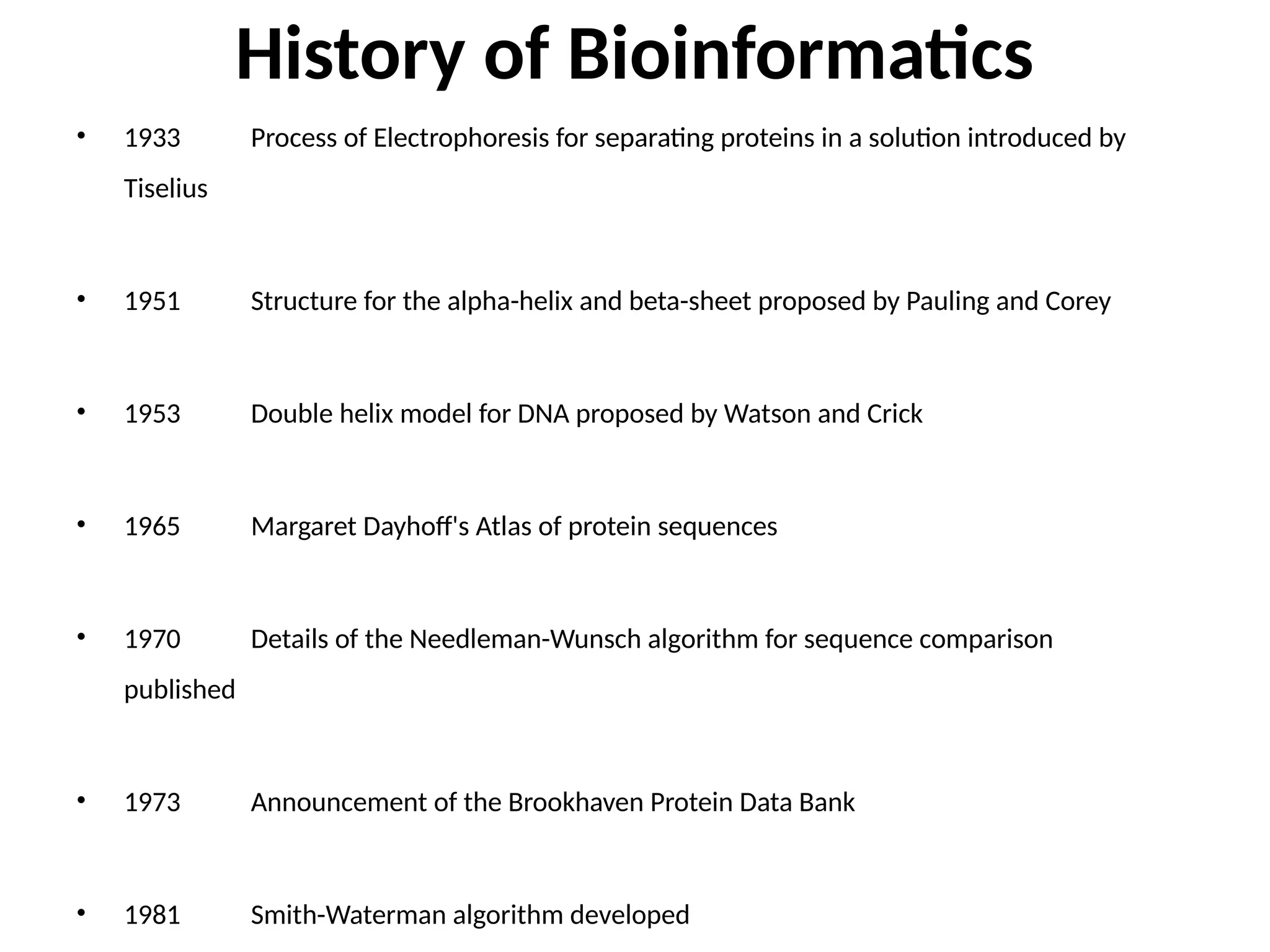
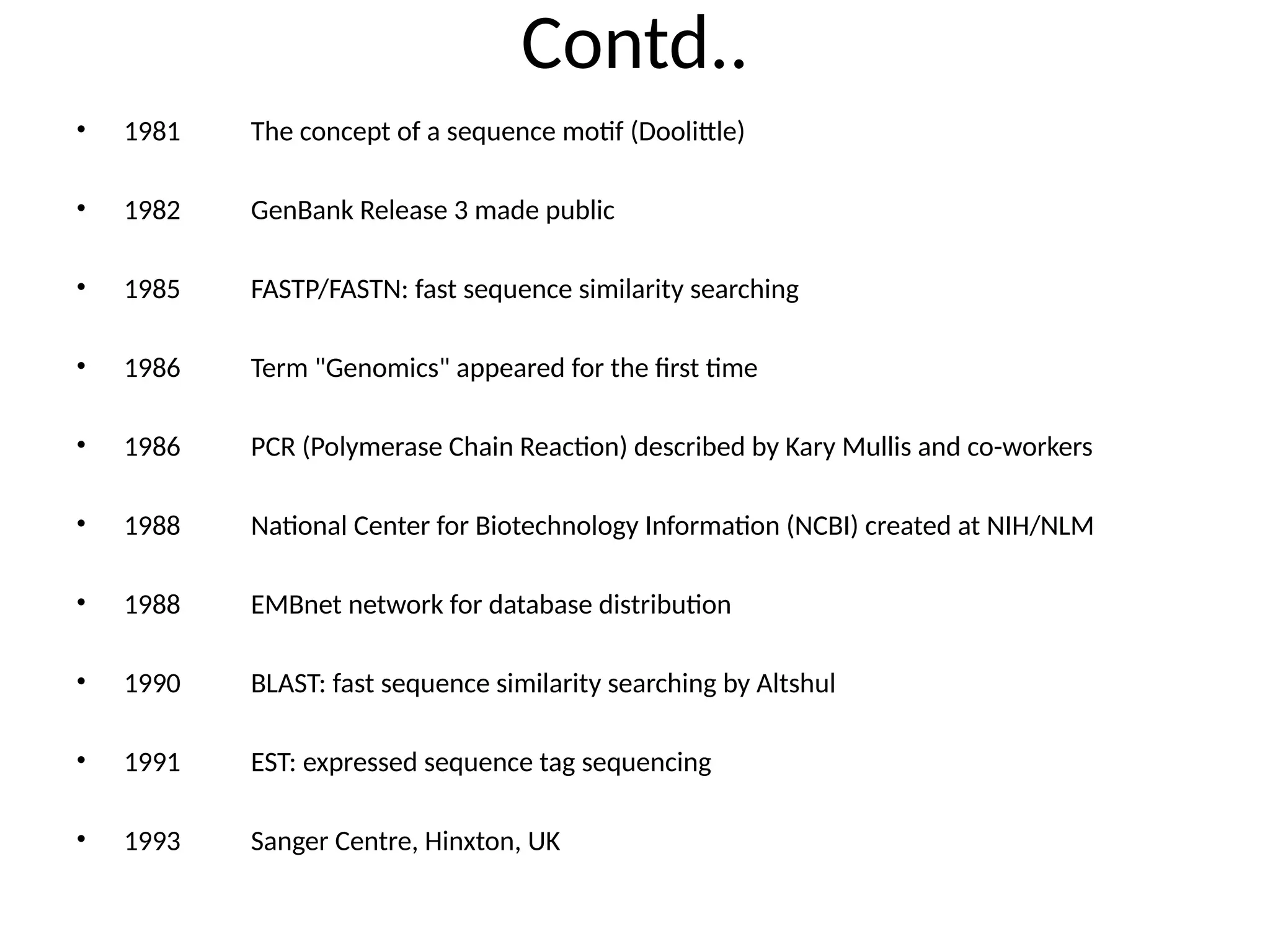
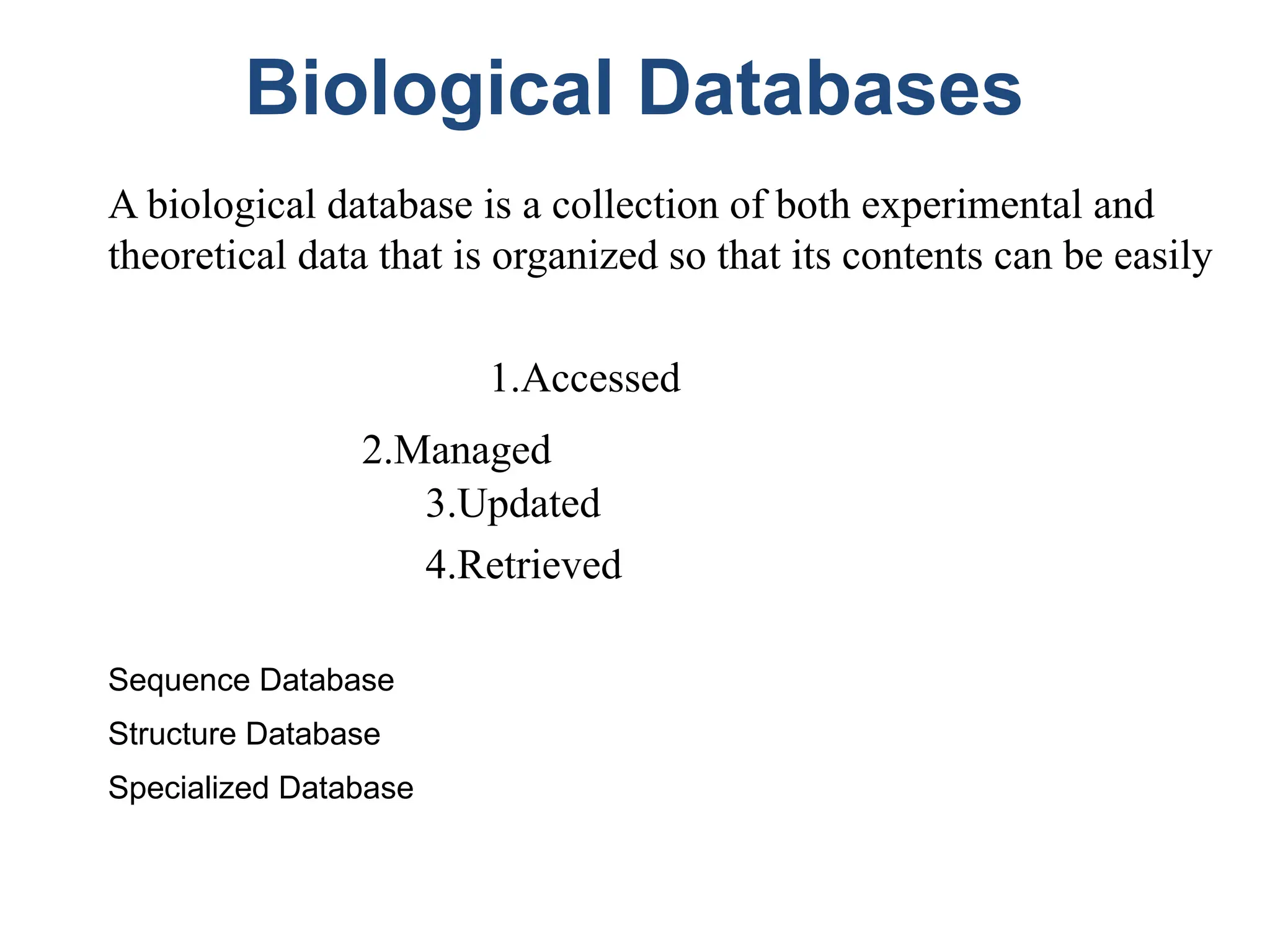
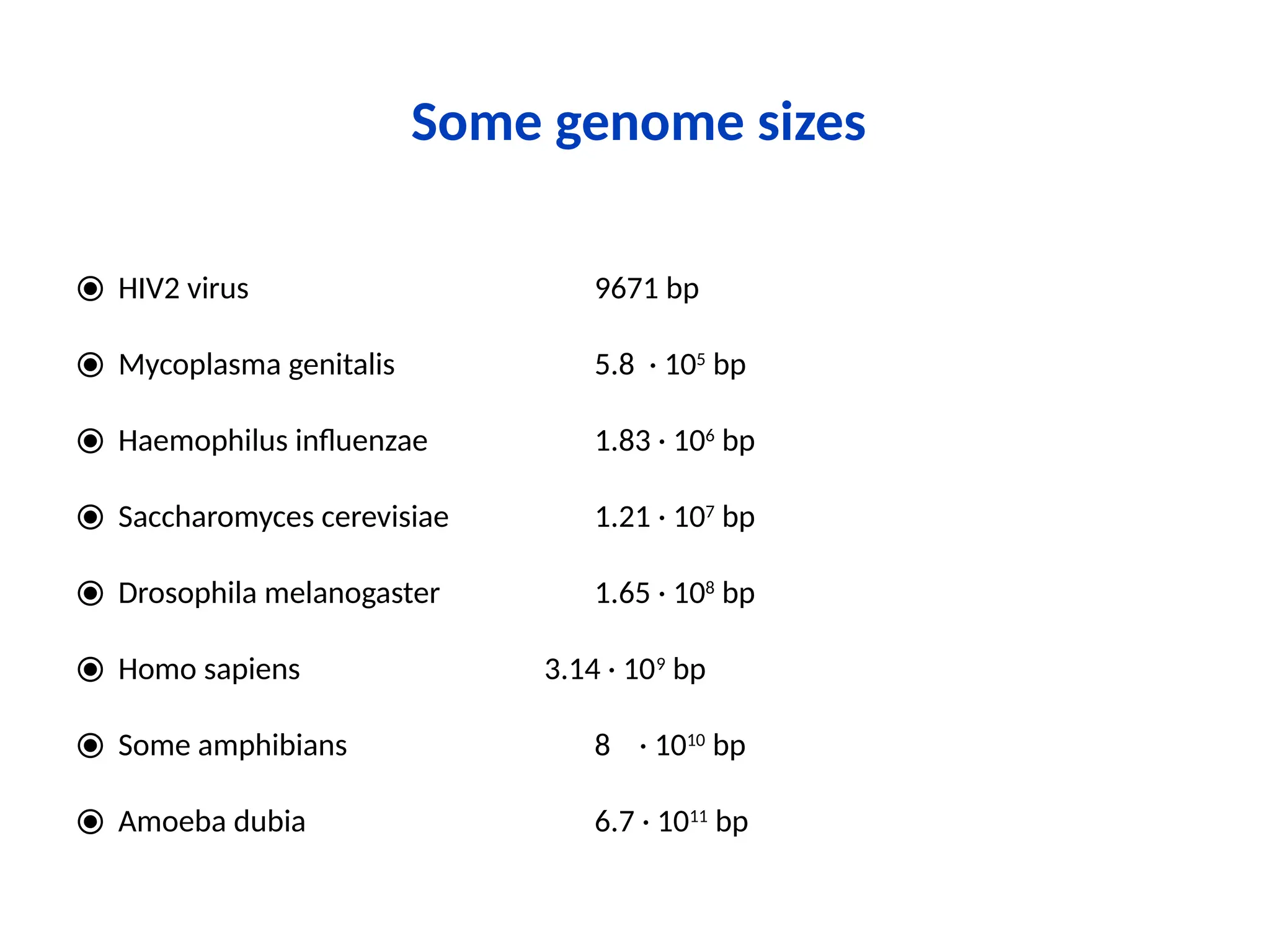
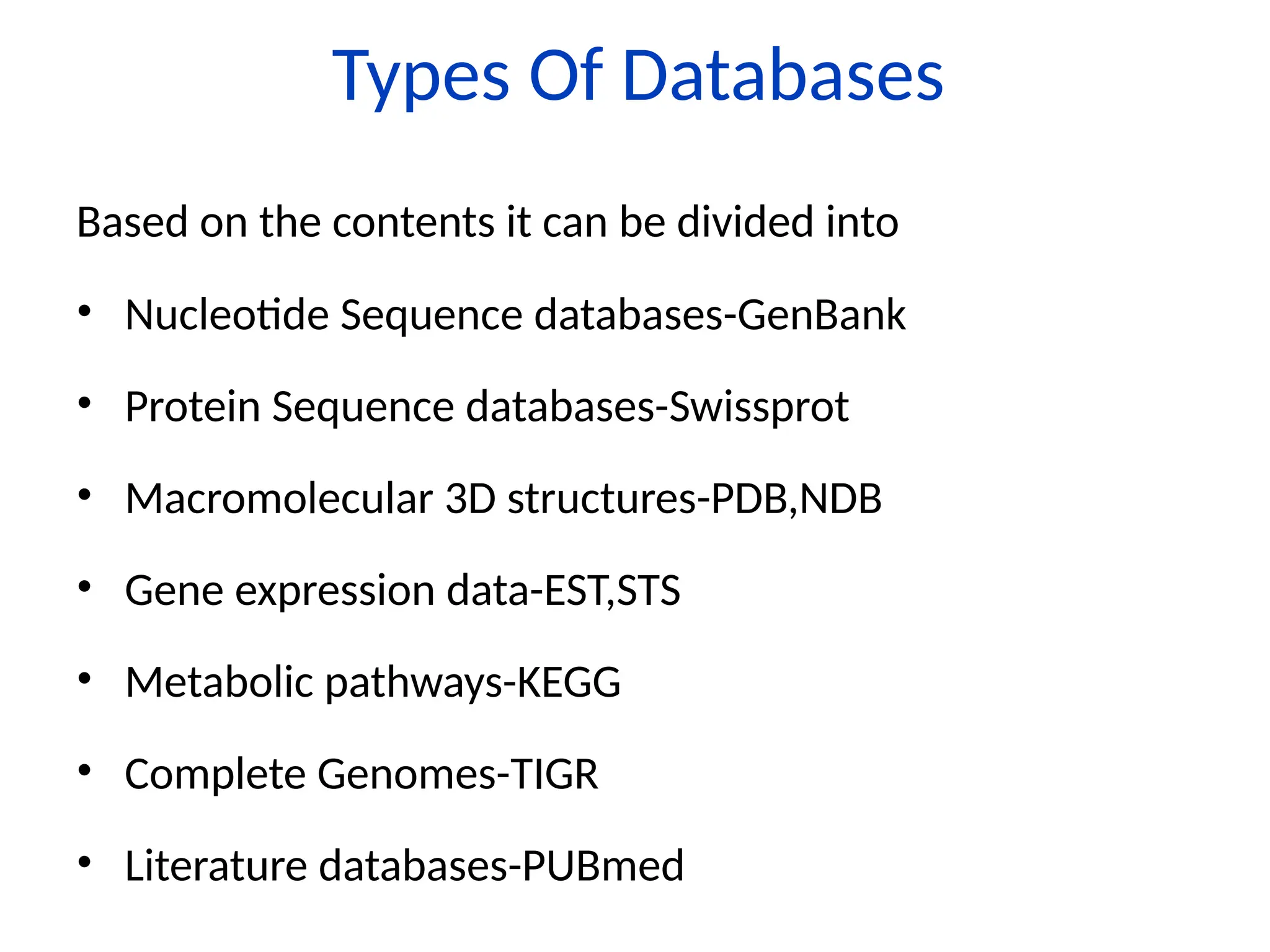
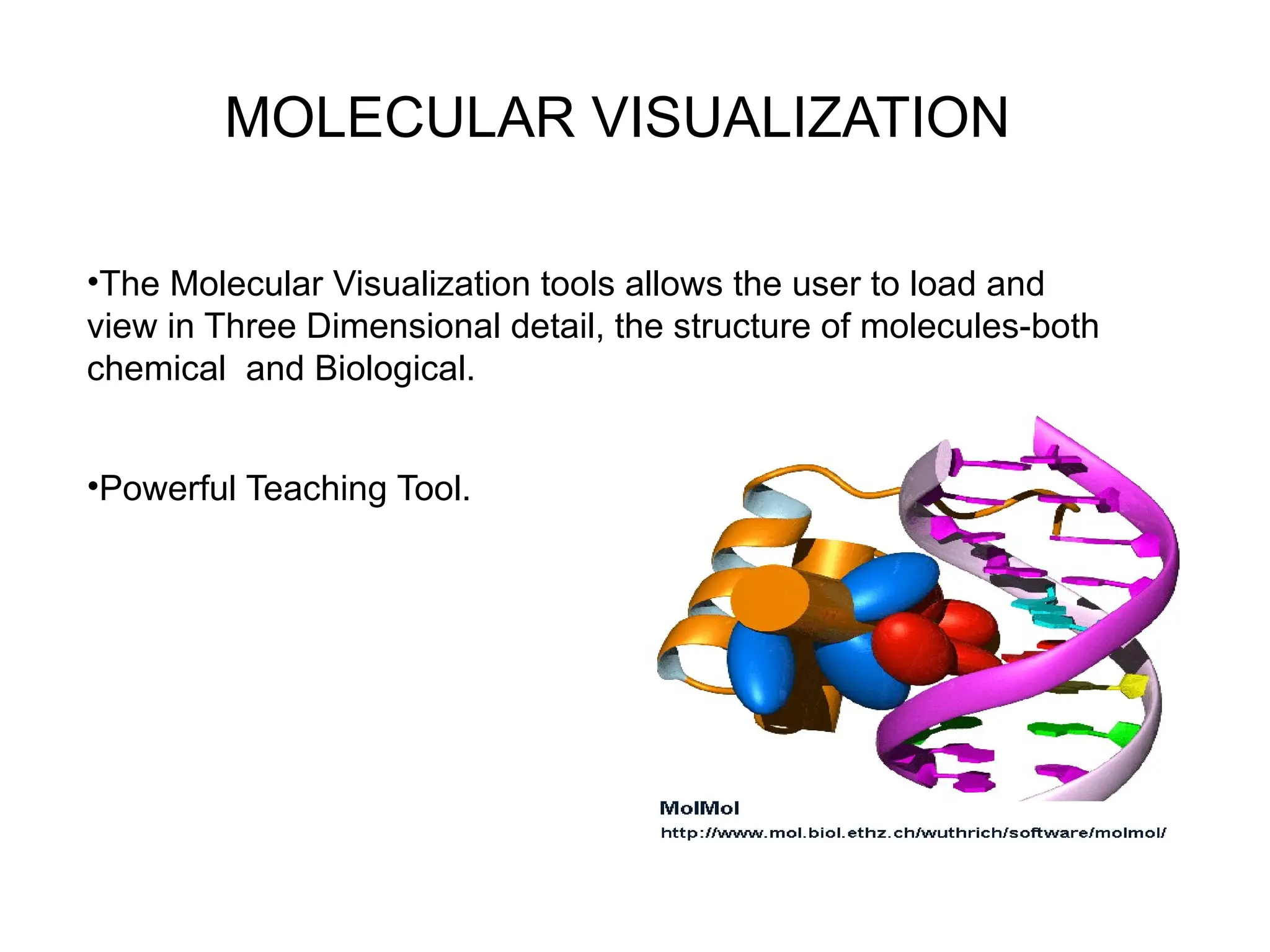
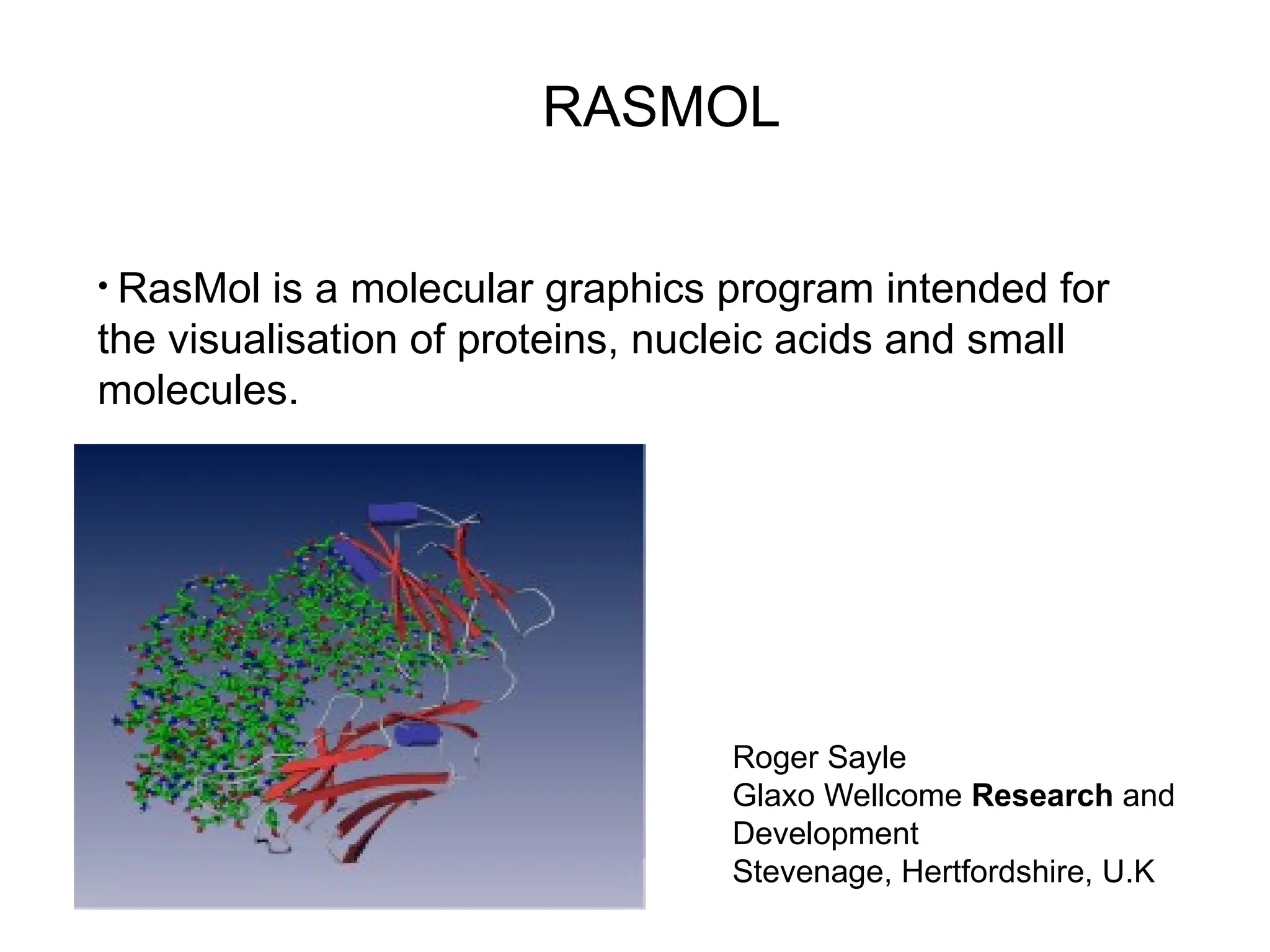
Bioinformatics is the integration of biology, computer science, and information technology focused on managing and analyzing biological data, particularly DNA and protein sequences. Its development has evolved alongside advancements in computing and molecular biology, with significant historical milestones from the 1930s onward, including key algorithms and the Human Genome Project. Bioinformatics encompasses various disciplines and applications, including genomics, proteomics, and molecular visualization, tackling challenges such as interdisciplinary collaboration and data dependency.