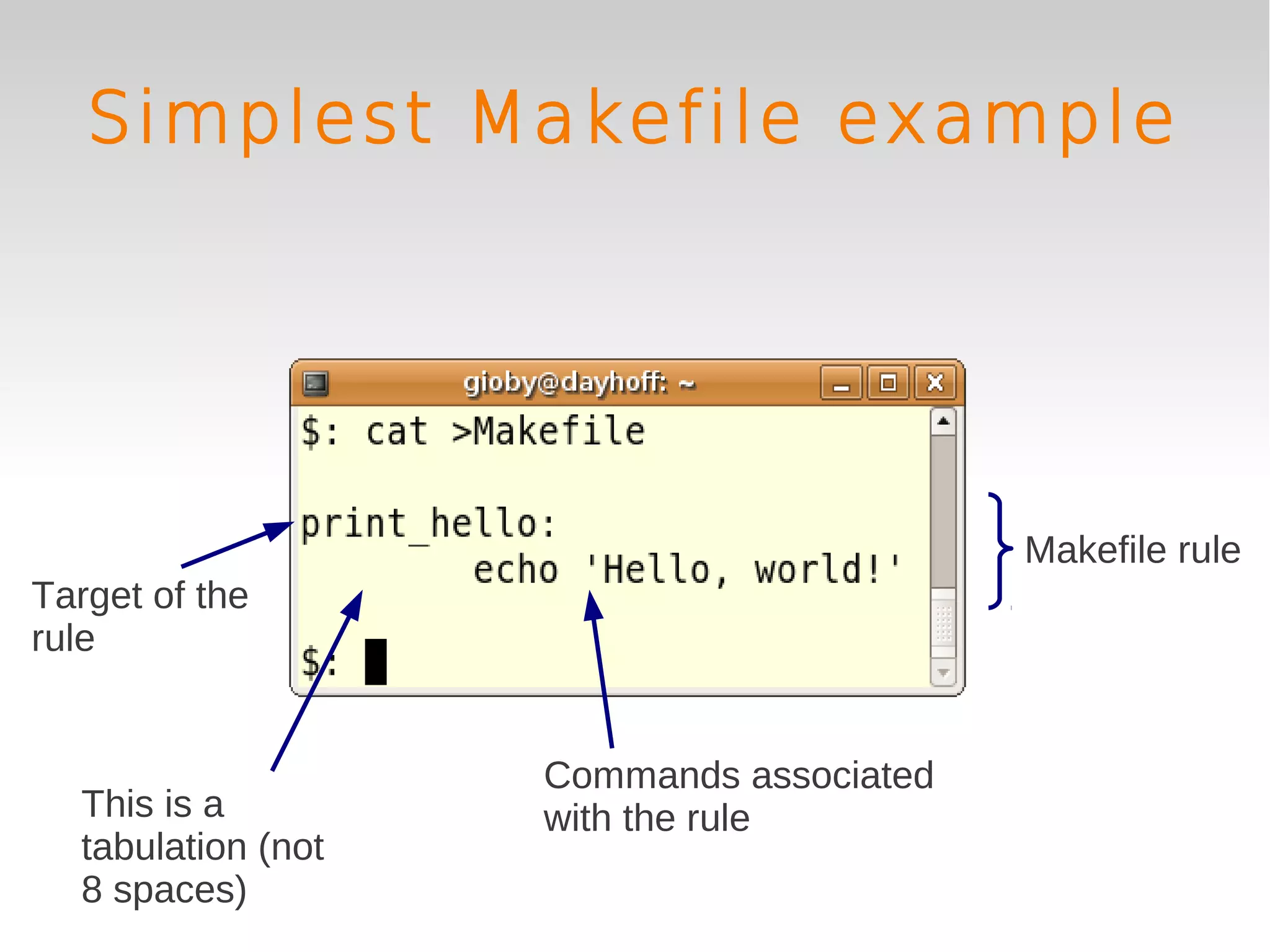
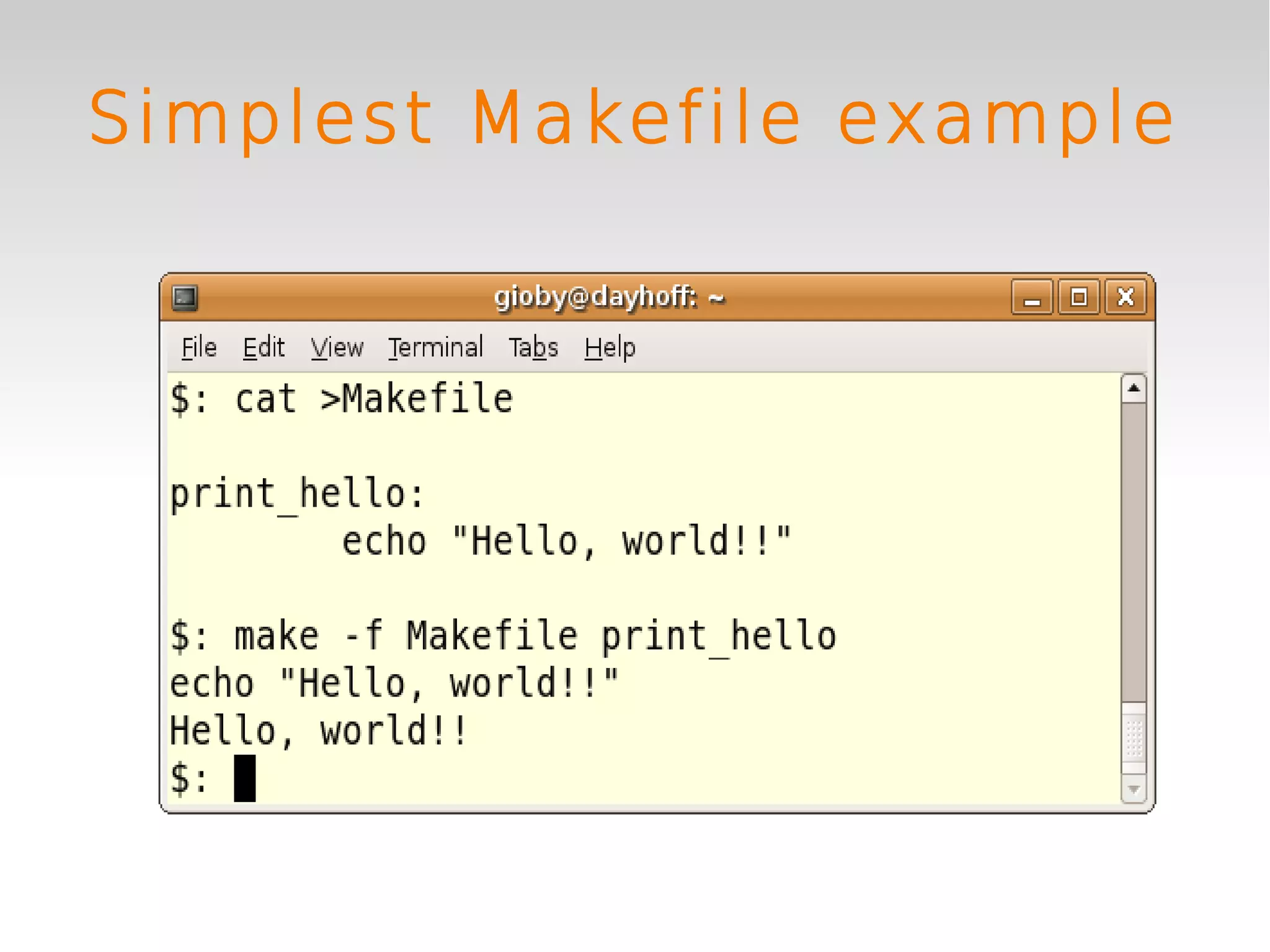
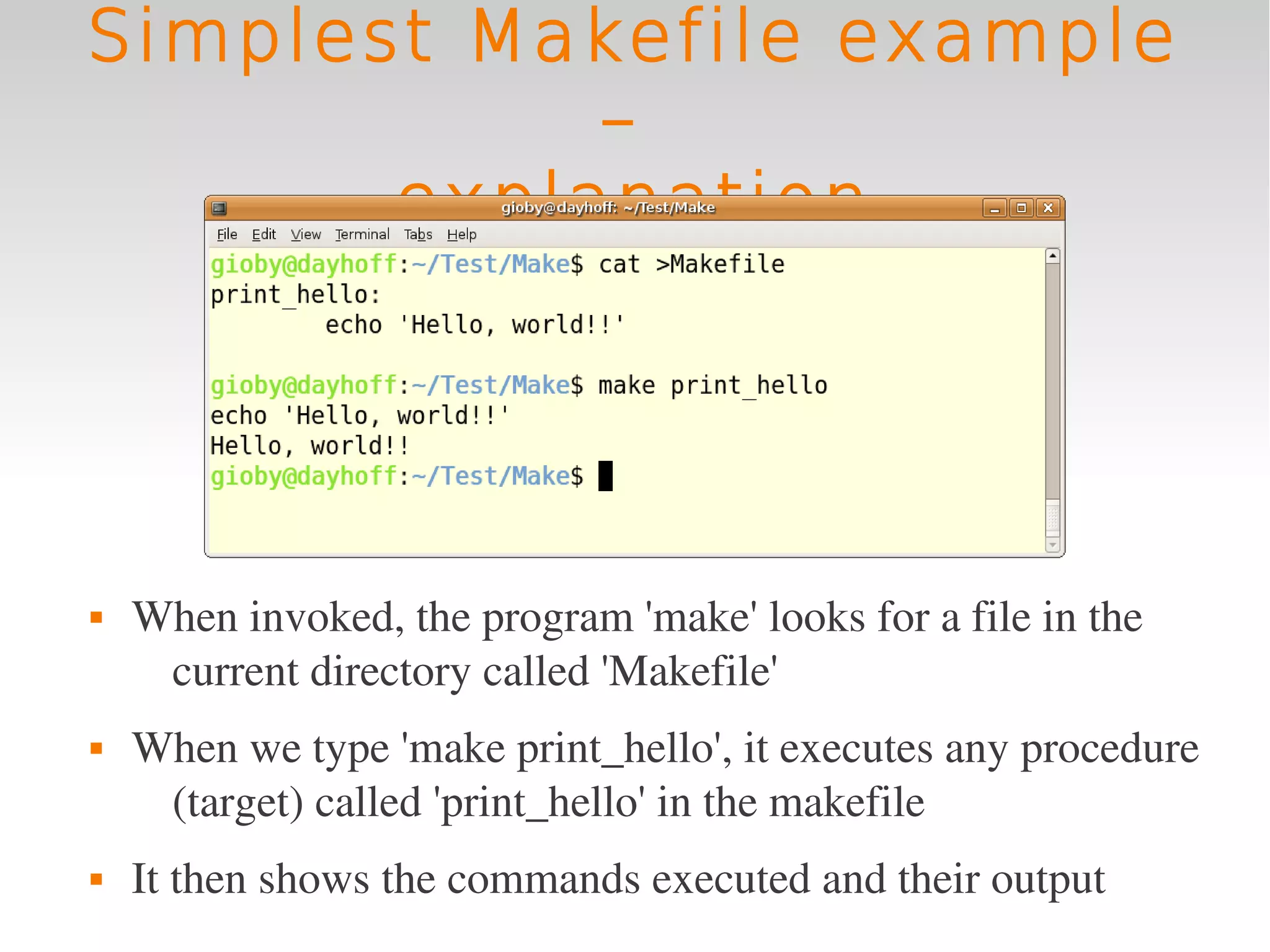
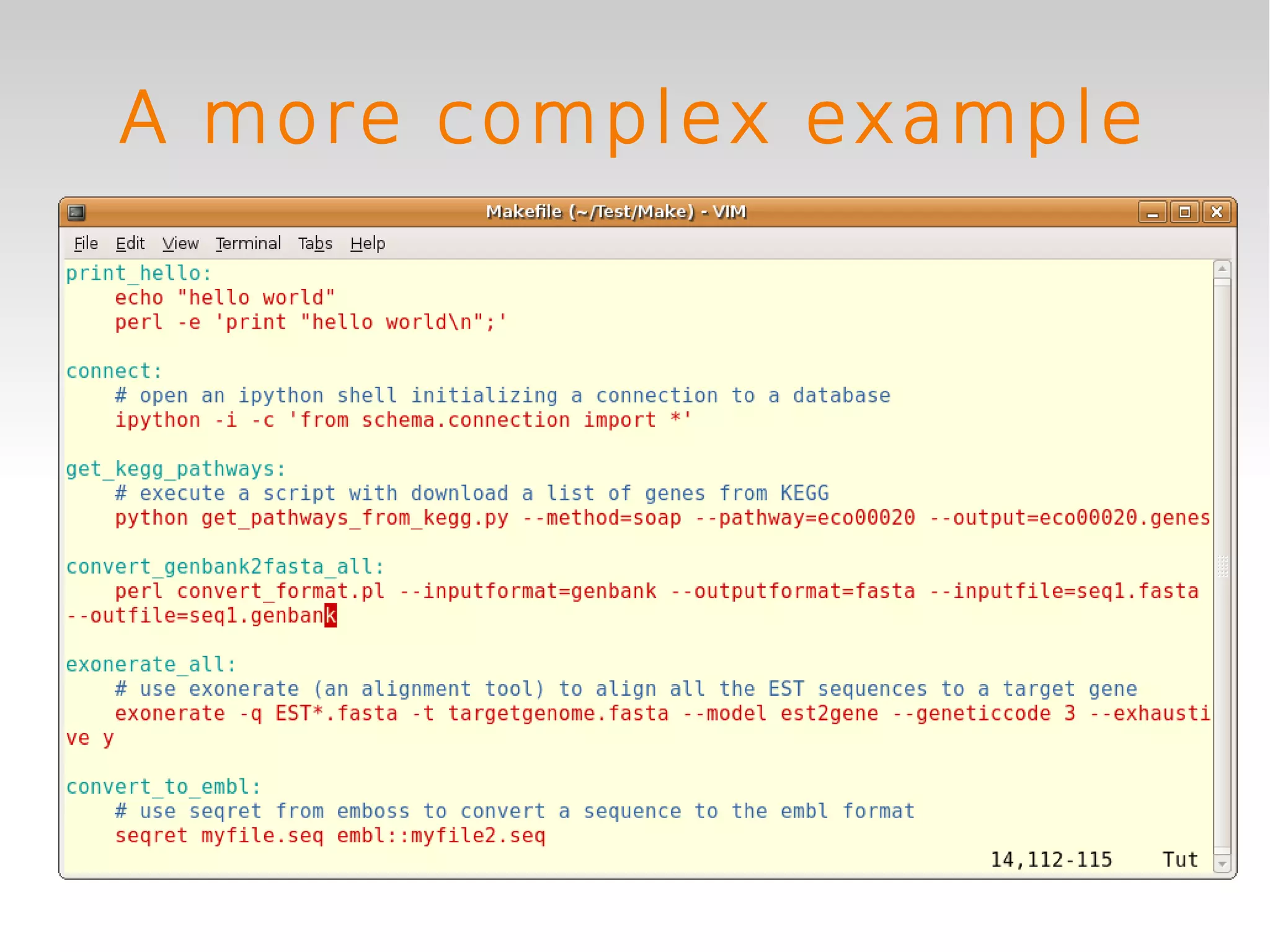
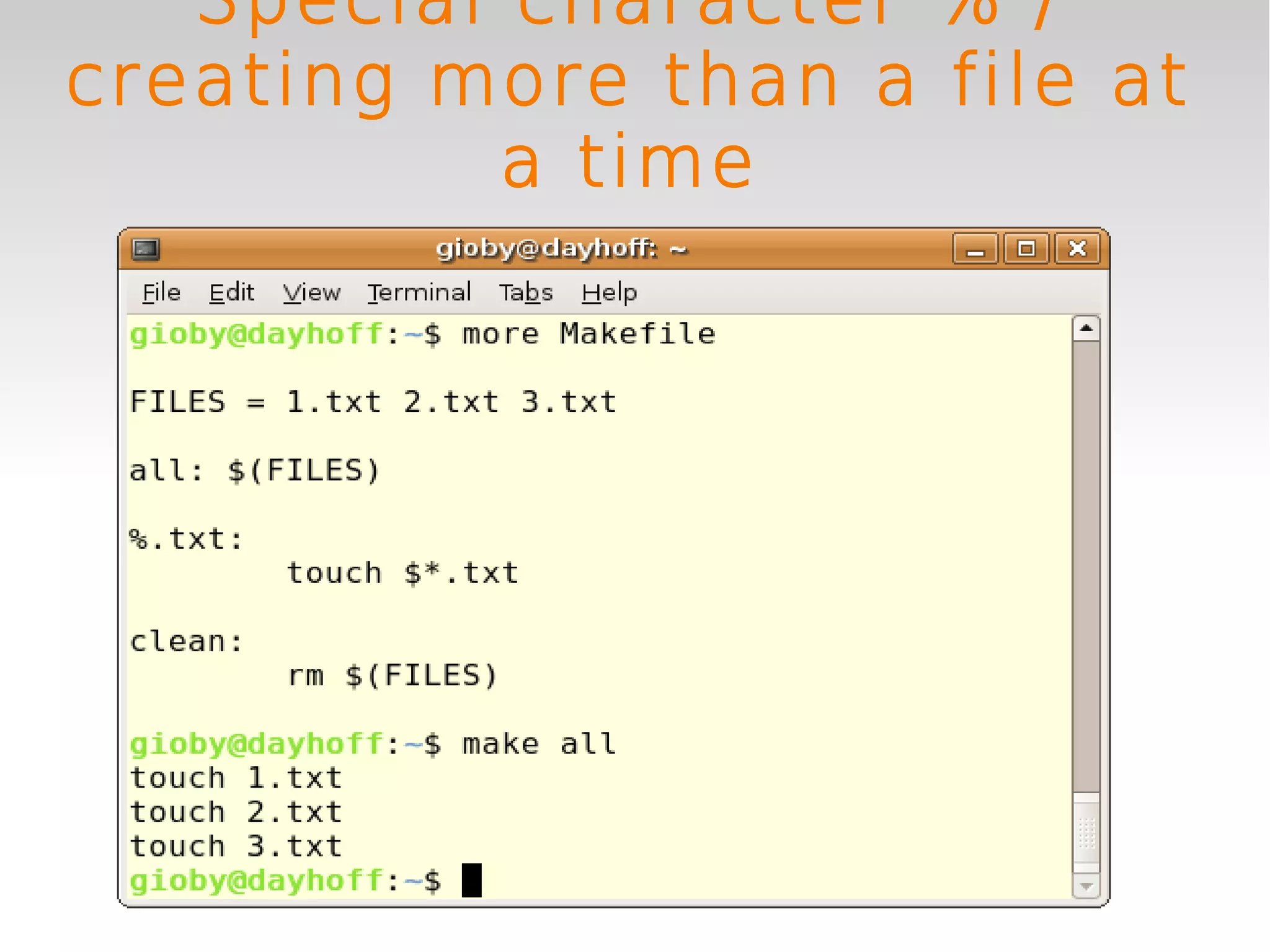
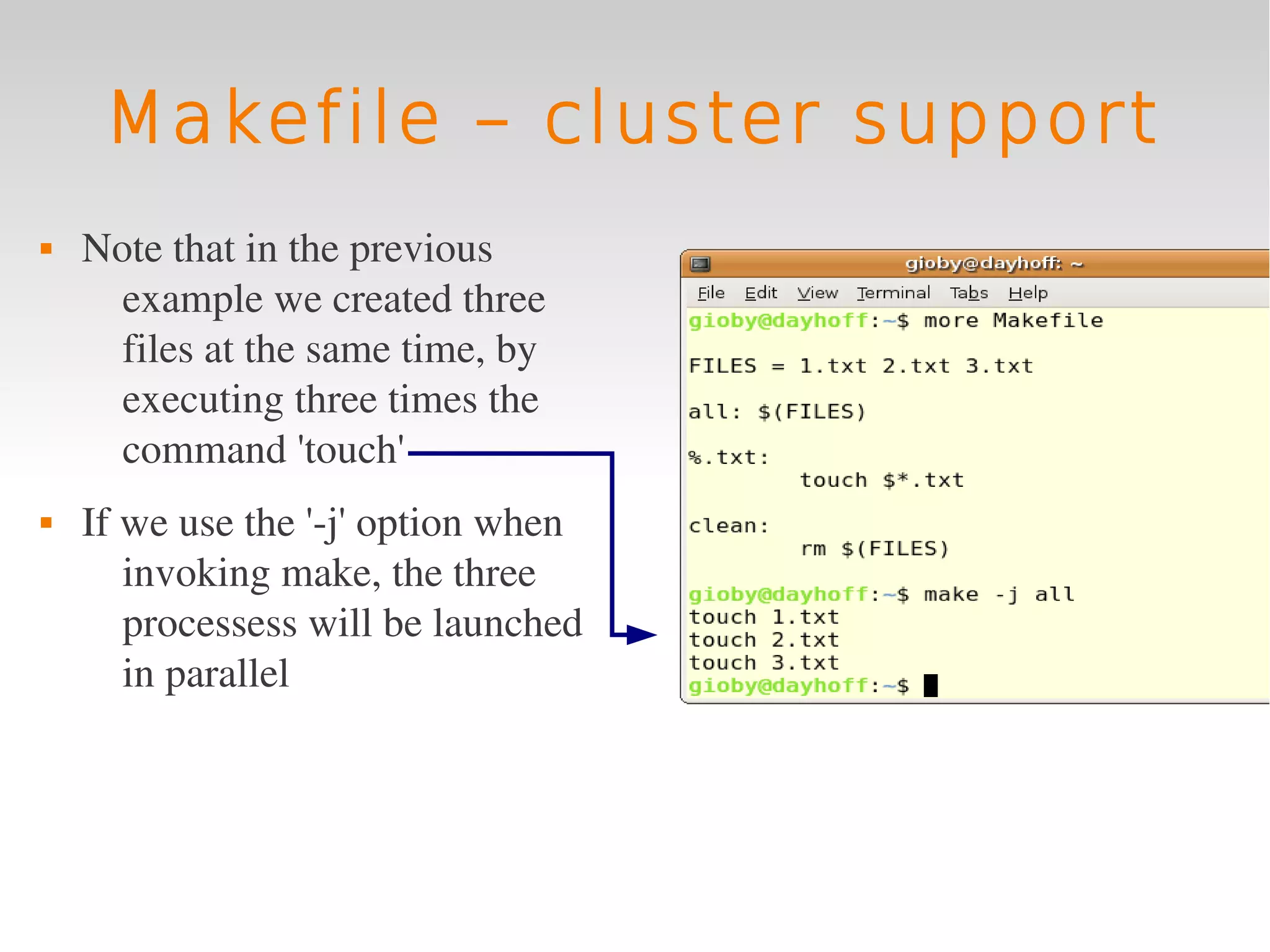
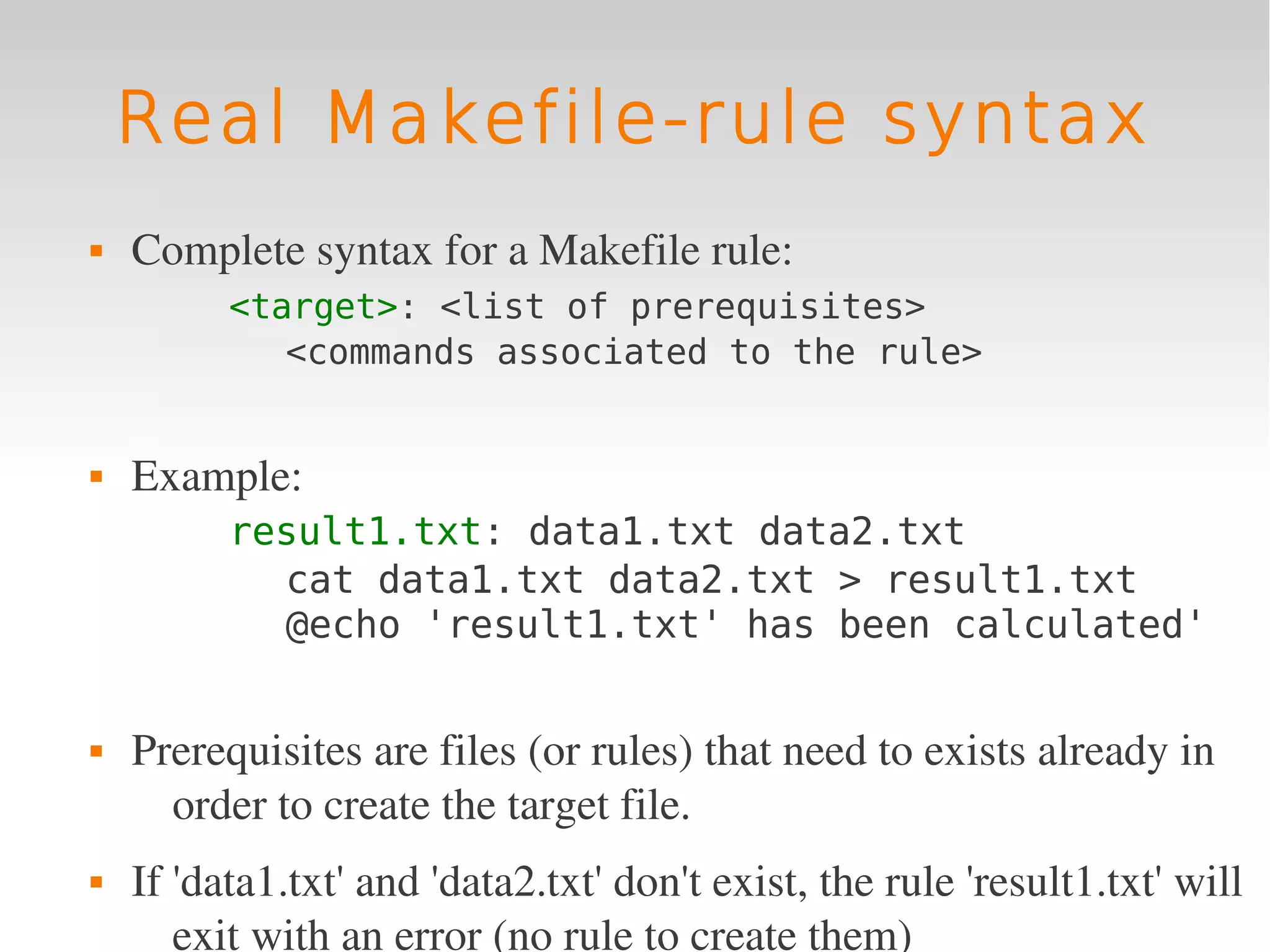
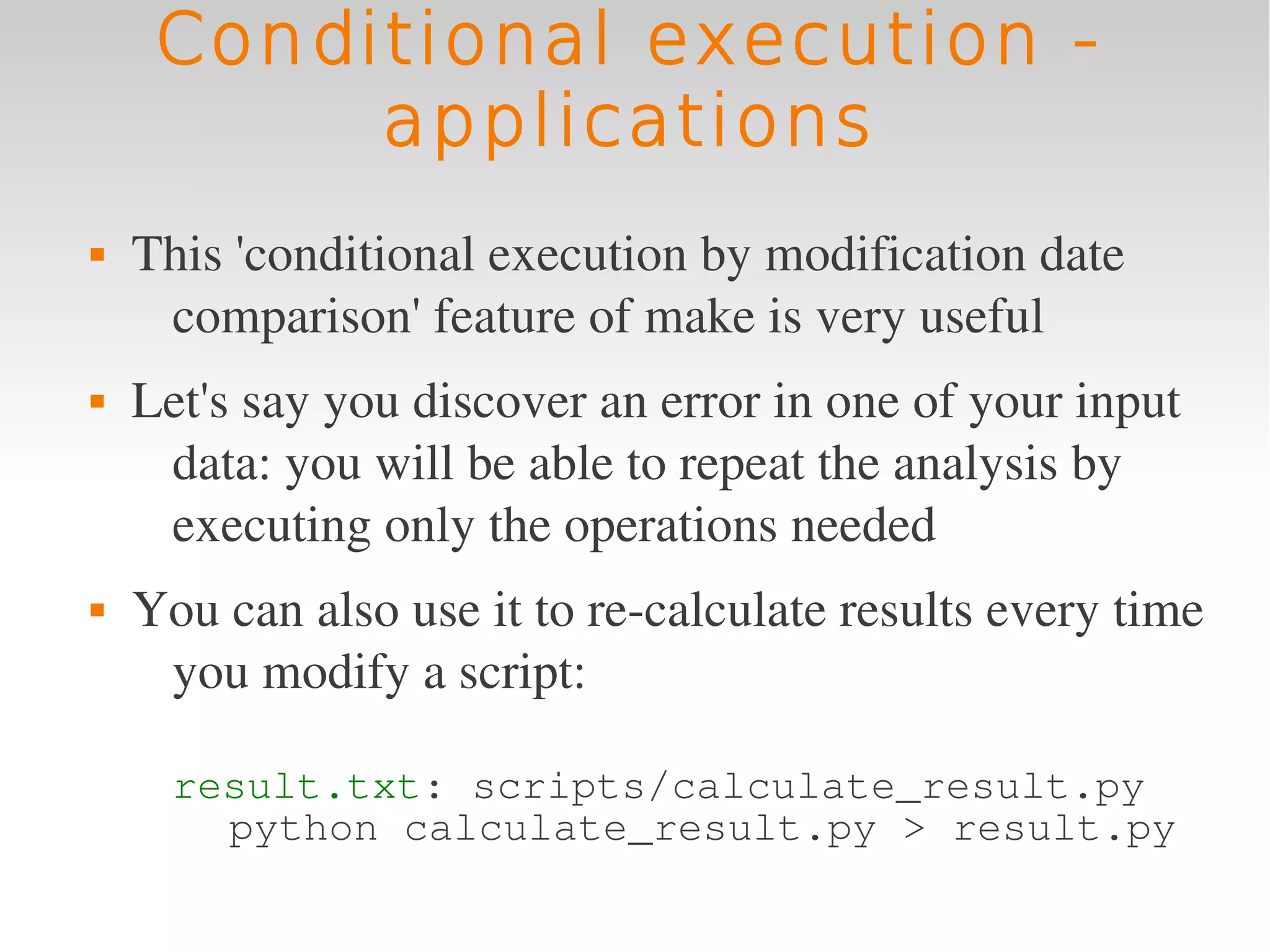
This document provides an introduction to using GNU Make, a tool for managing dependencies and automating command execution in Unix systems. It explains the basic syntax and functionality of makefiles, including creating tasks, handling prerequisites, and using variables. The document also covers advanced features such as conditional execution, parallel processing, and alternatives to Make in bioinformatics.