Downloaded 296 times

















The document presents the procksi-server, an online decision support system designed for protein structure comparison. It discusses the importance of protein structures, existing comparison methods, and introduces procksi's unique approaches, including its home-grown methods like universal similarity metric (USM) and maximum contact map overlap (max-CMO). Finally, it emphasizes the need for a comprehensive platform that integrates various methods and facilitates user-friendly analysis for protein structure researchers.


































































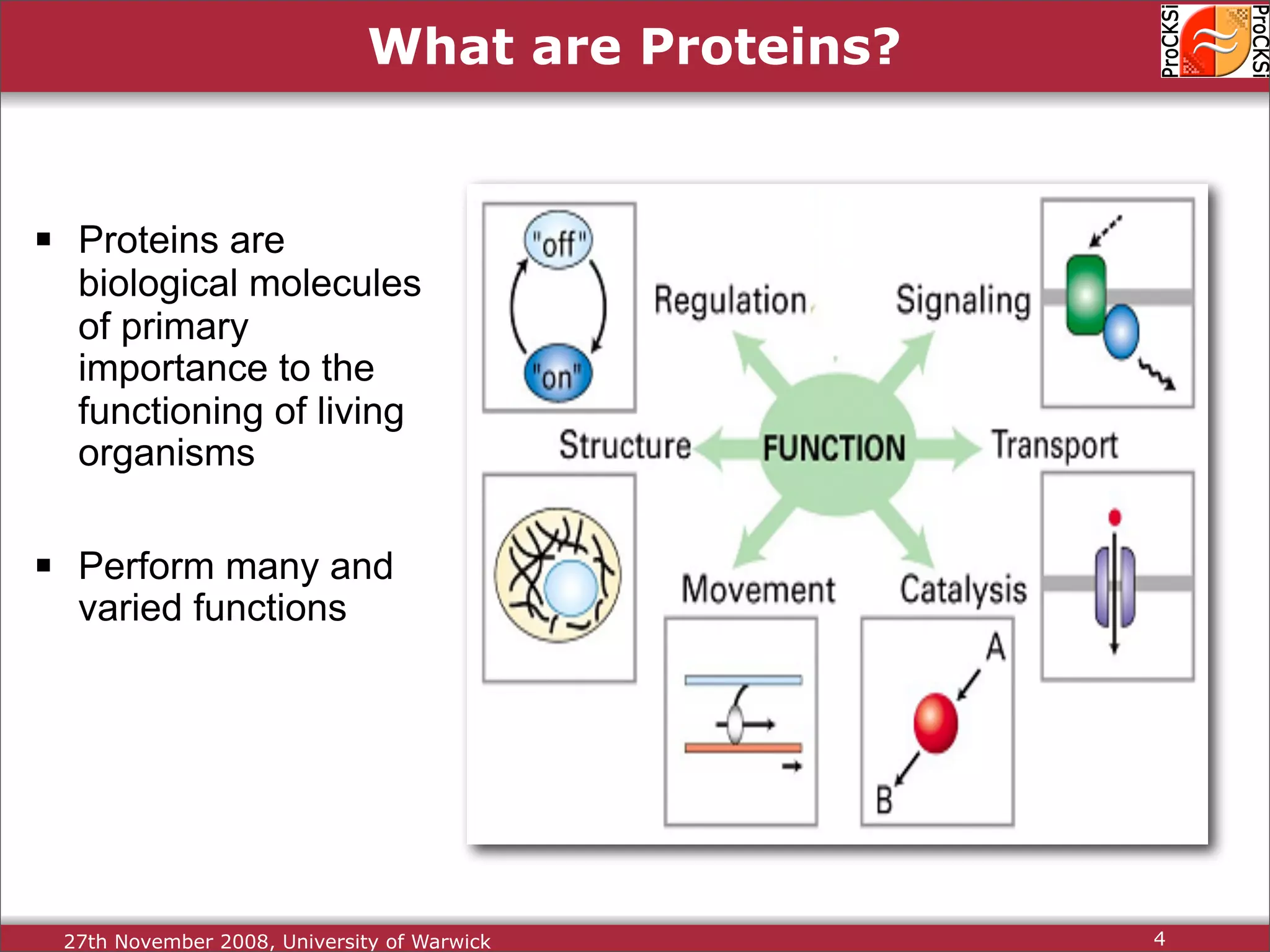
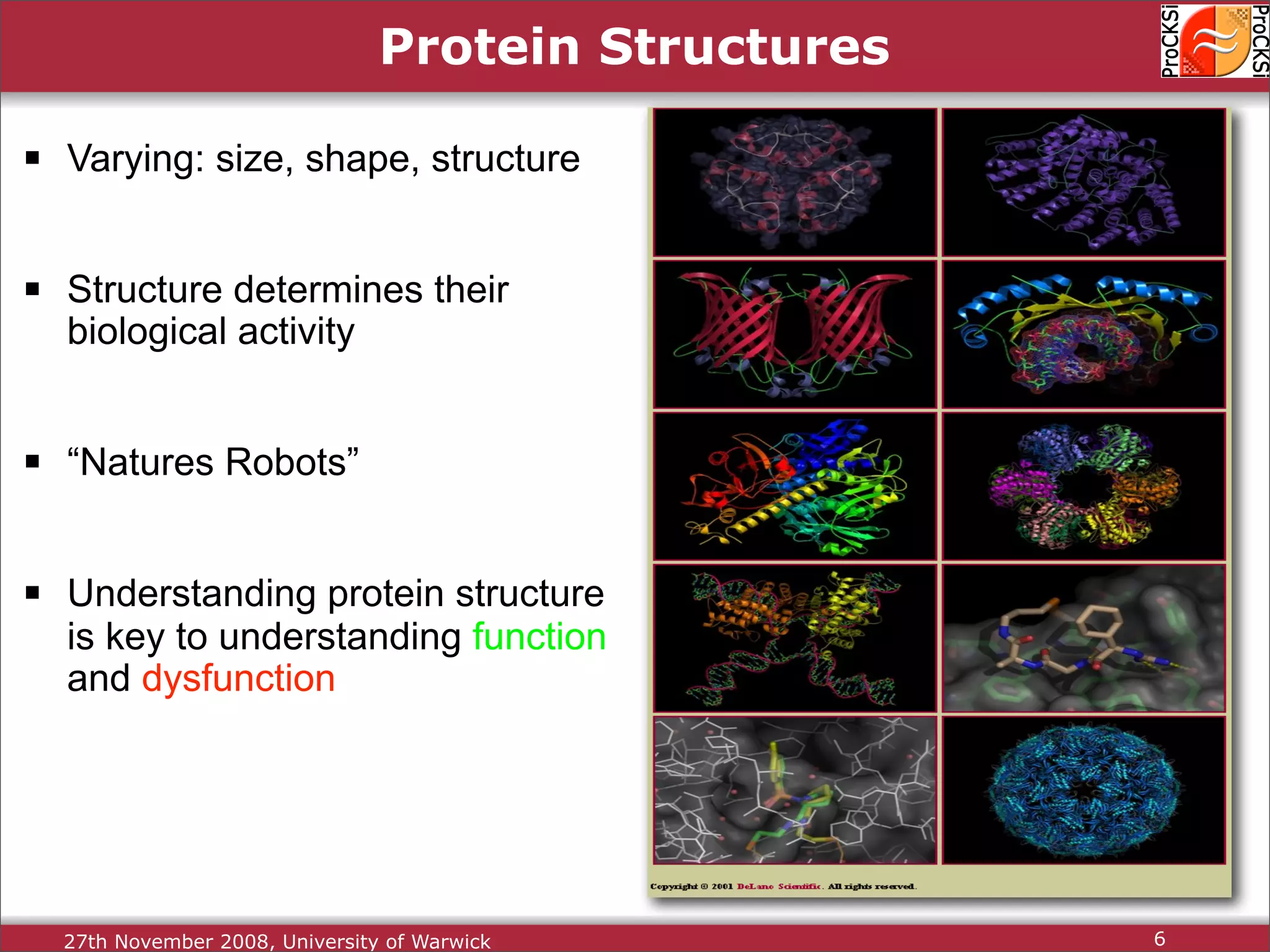
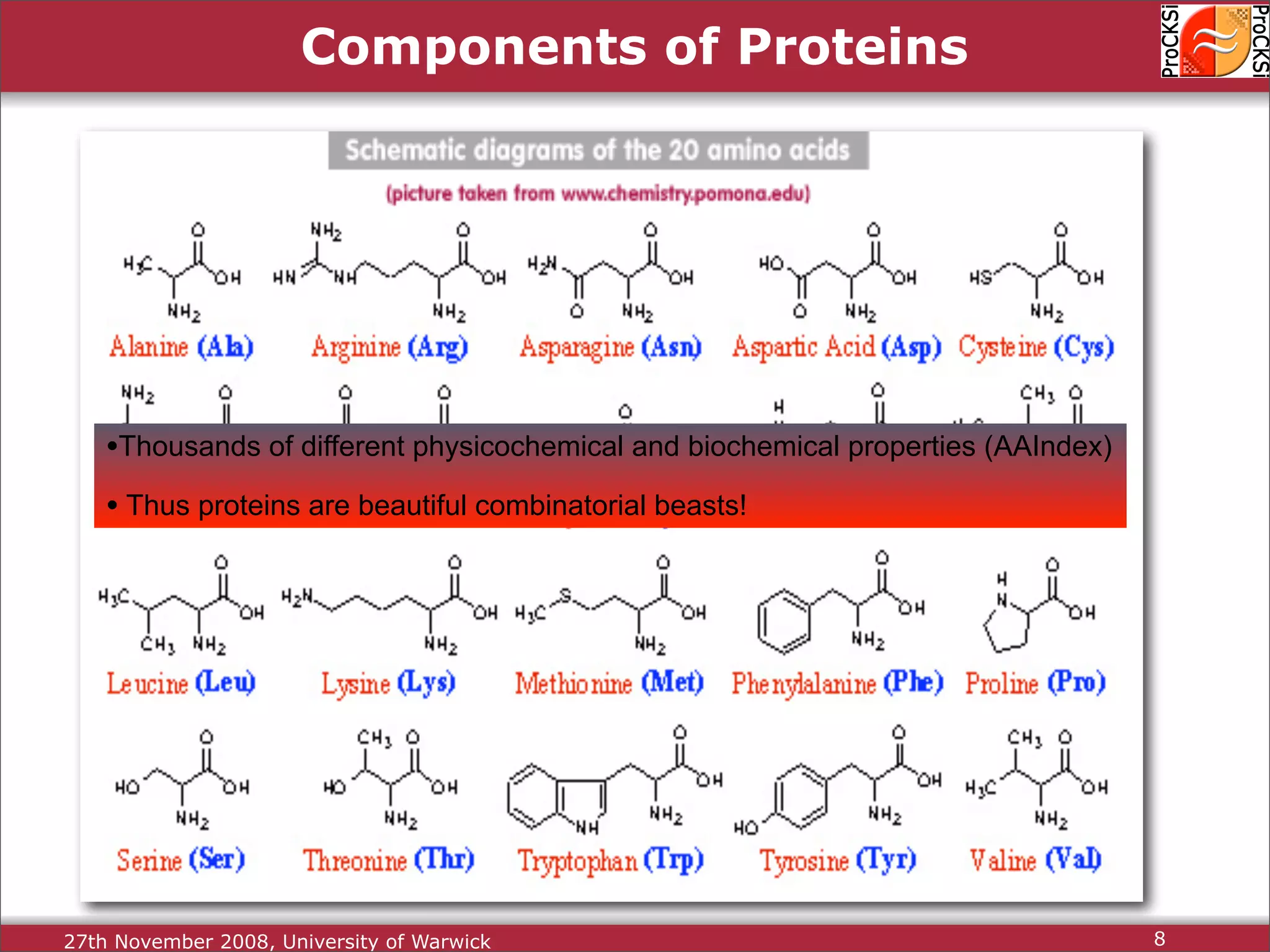
Overview of proteins, their functions, and the importance of understanding their structures.
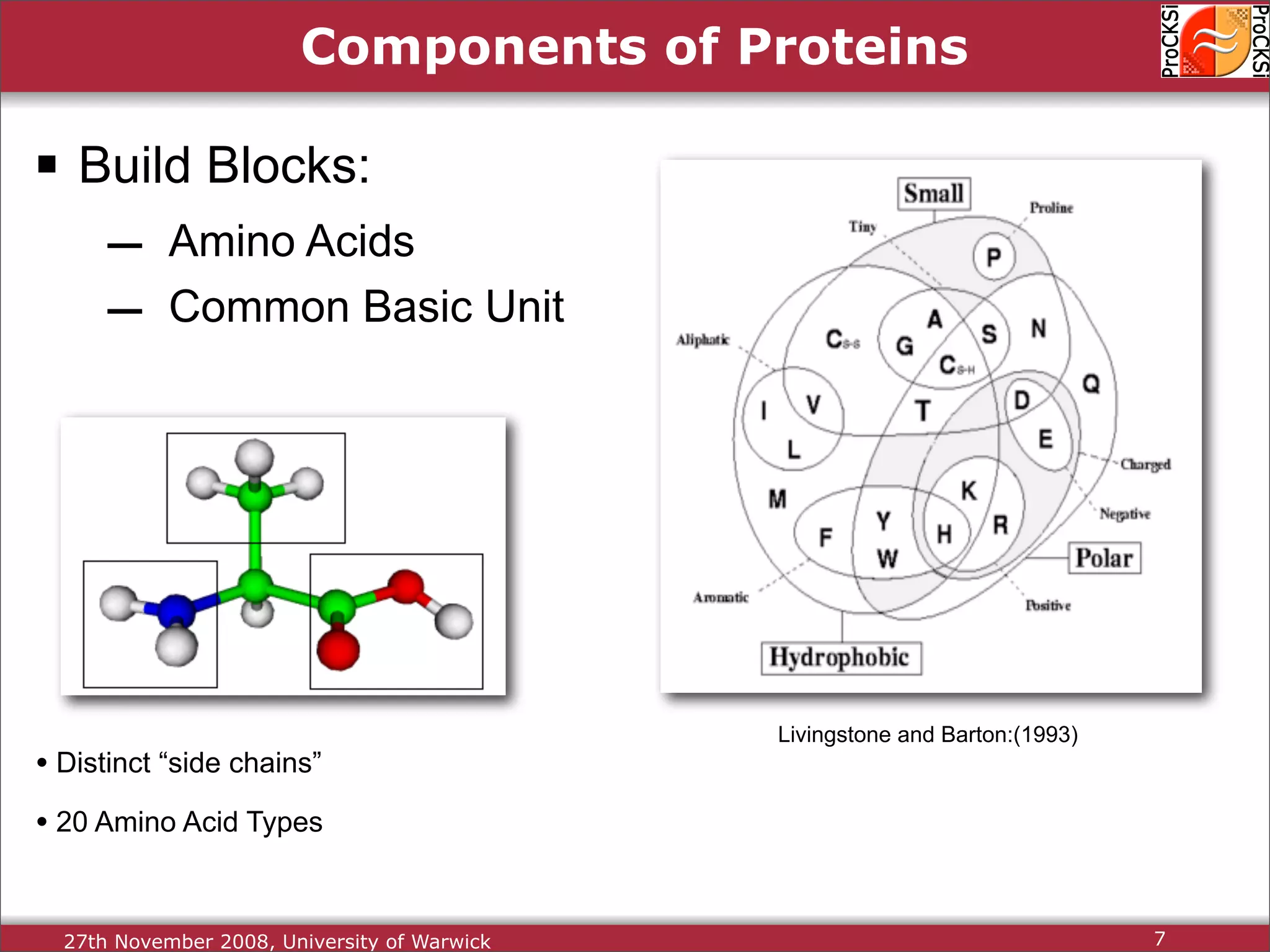
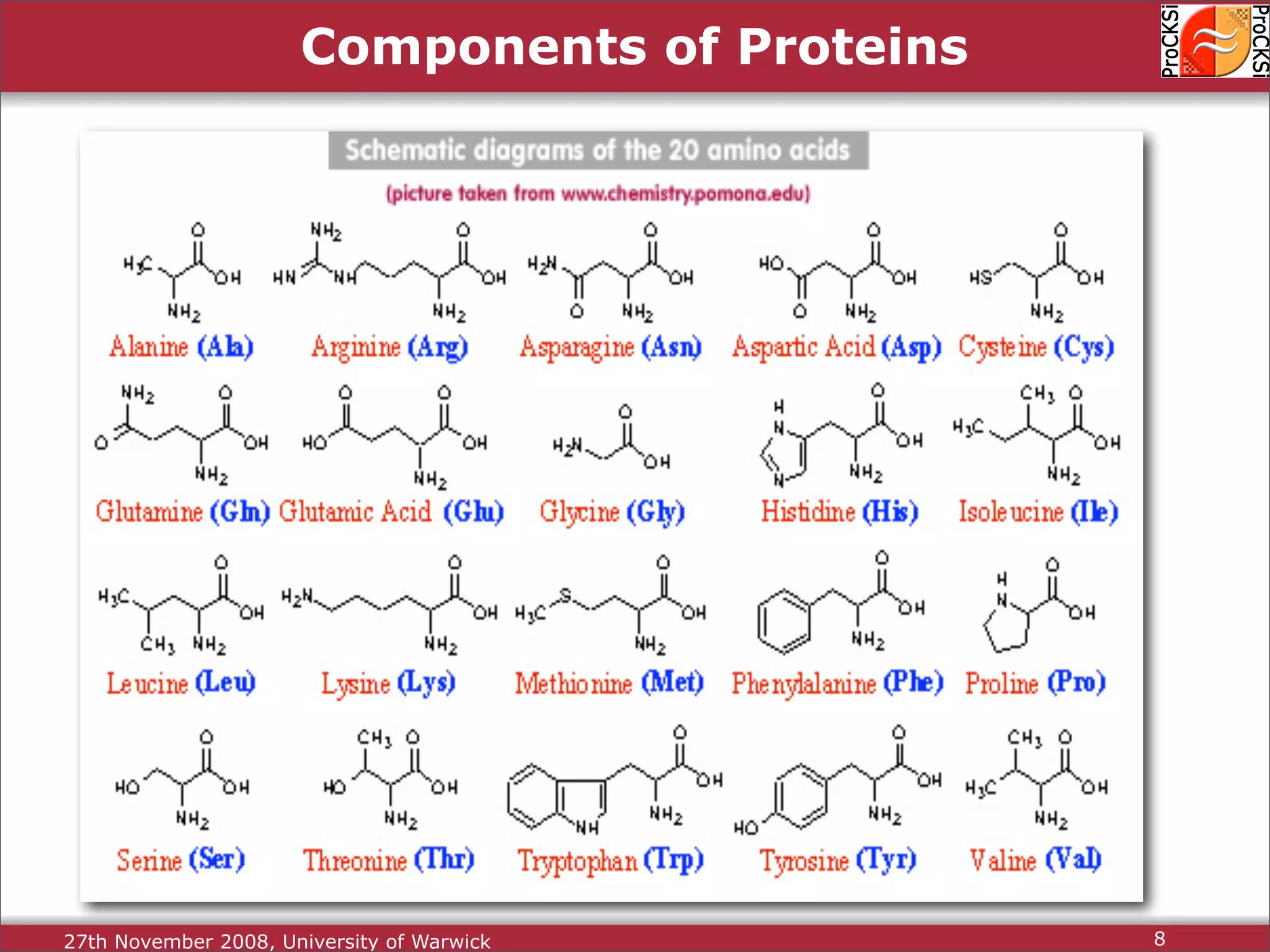
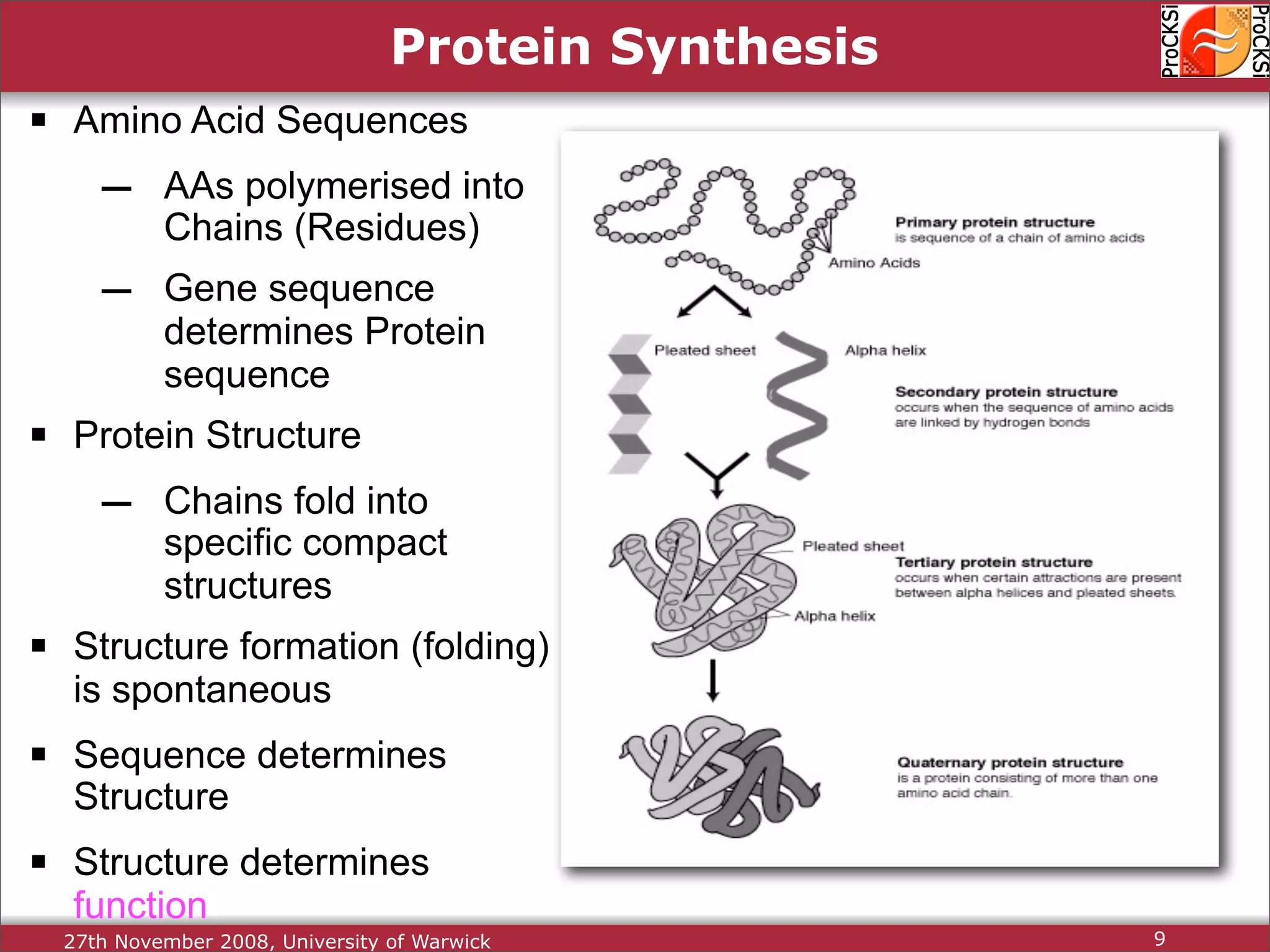
Discussion of amino acids, protein synthesis, and how sequences dictate structure and function.
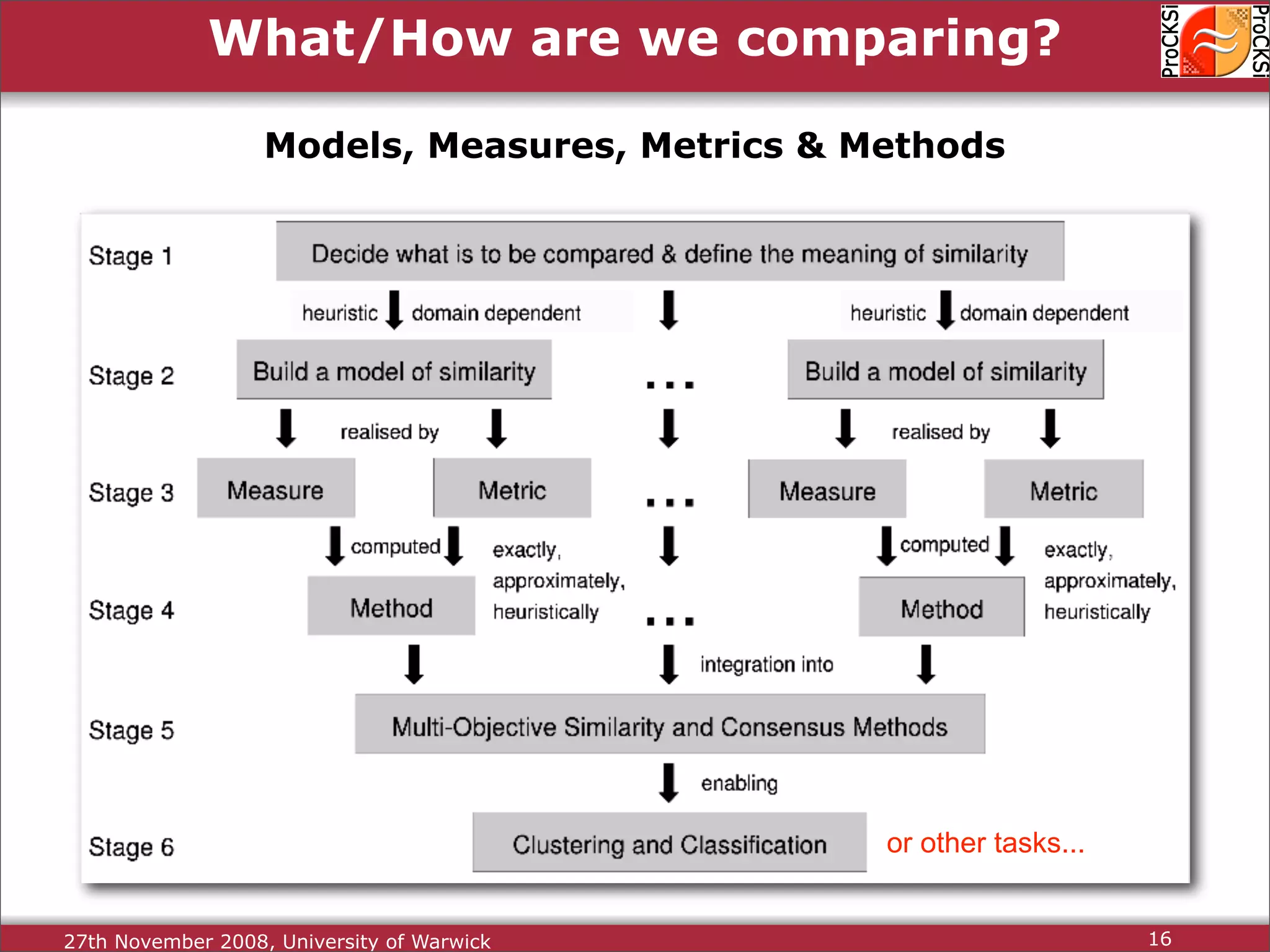
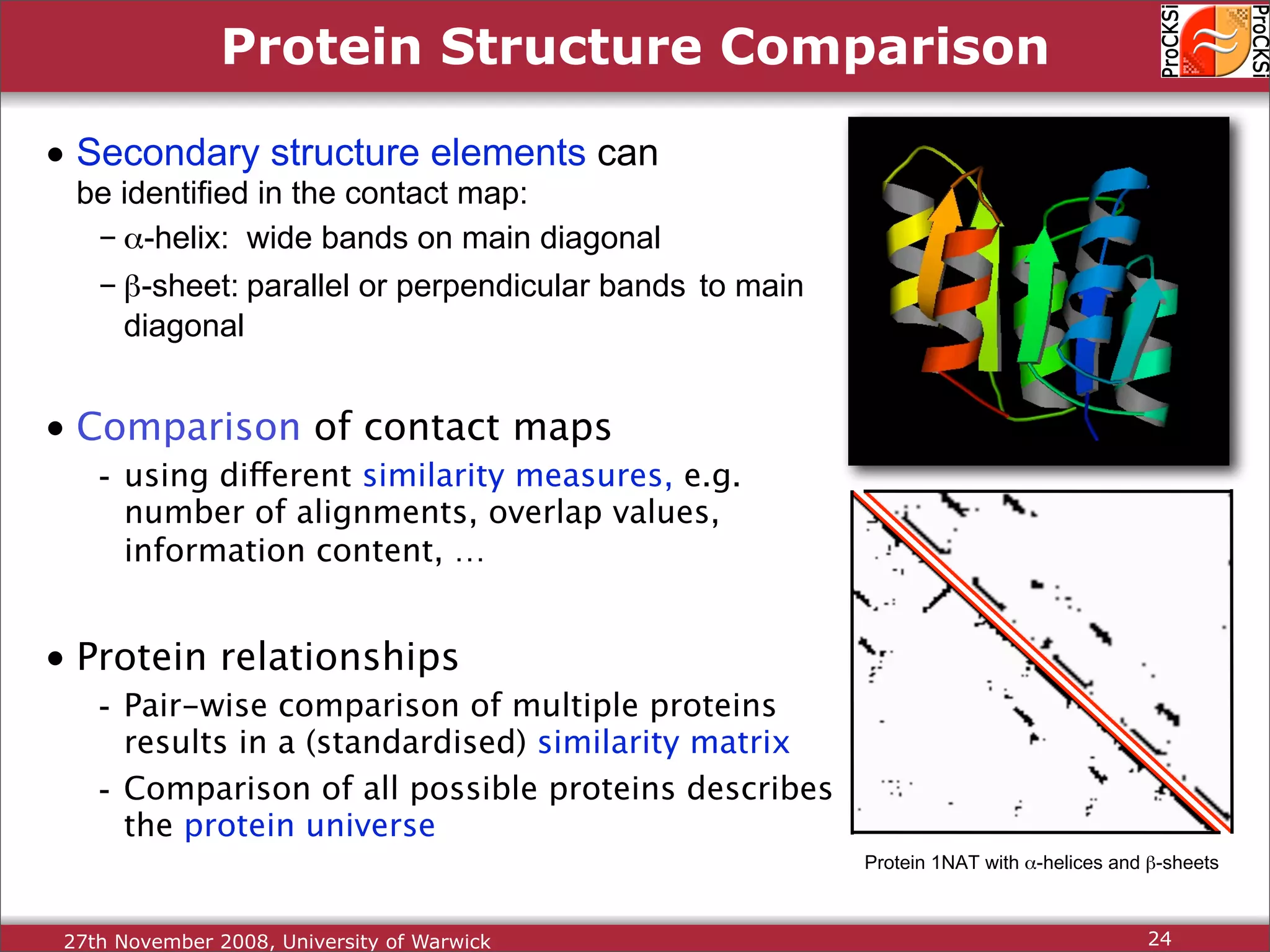
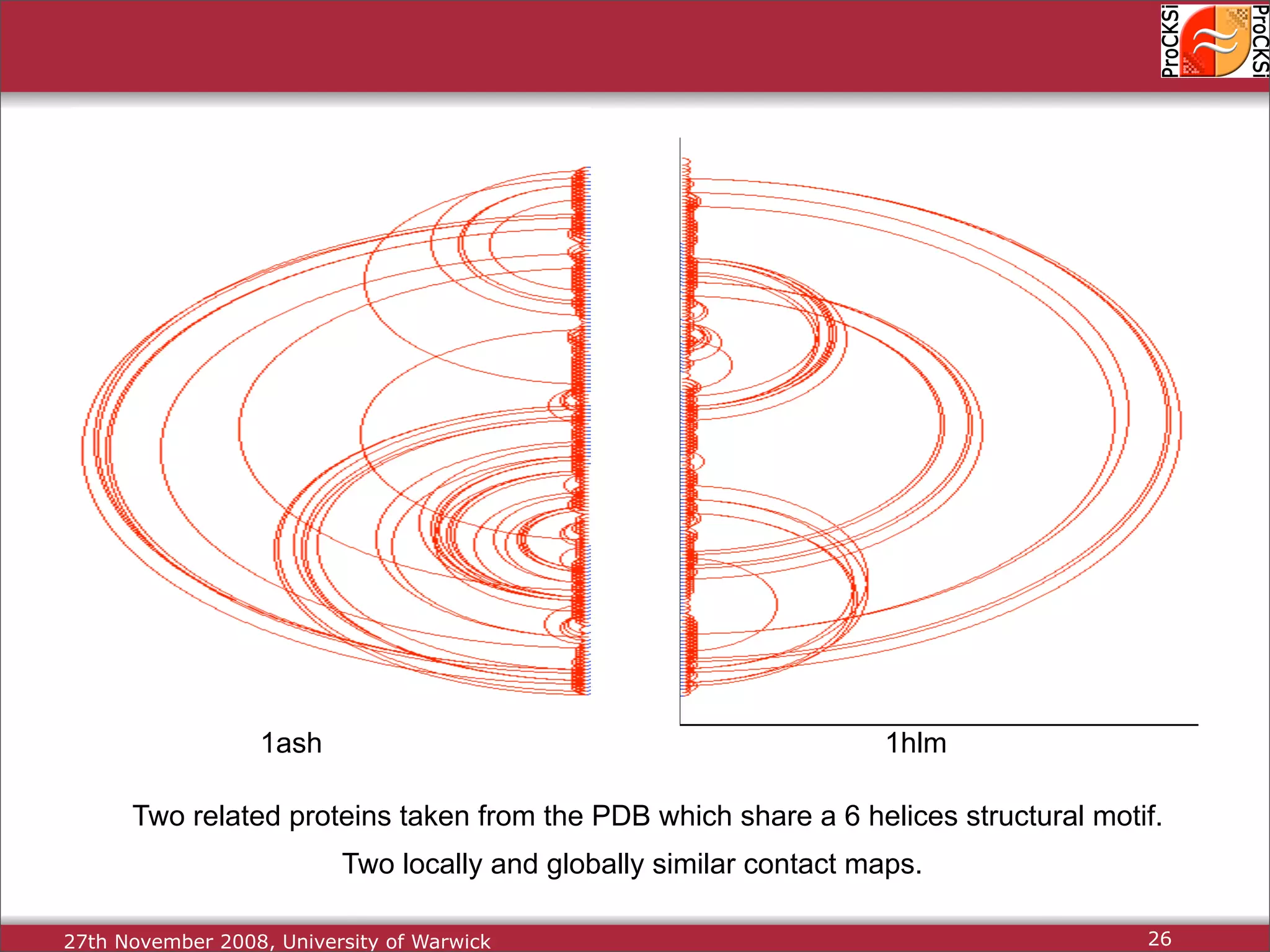
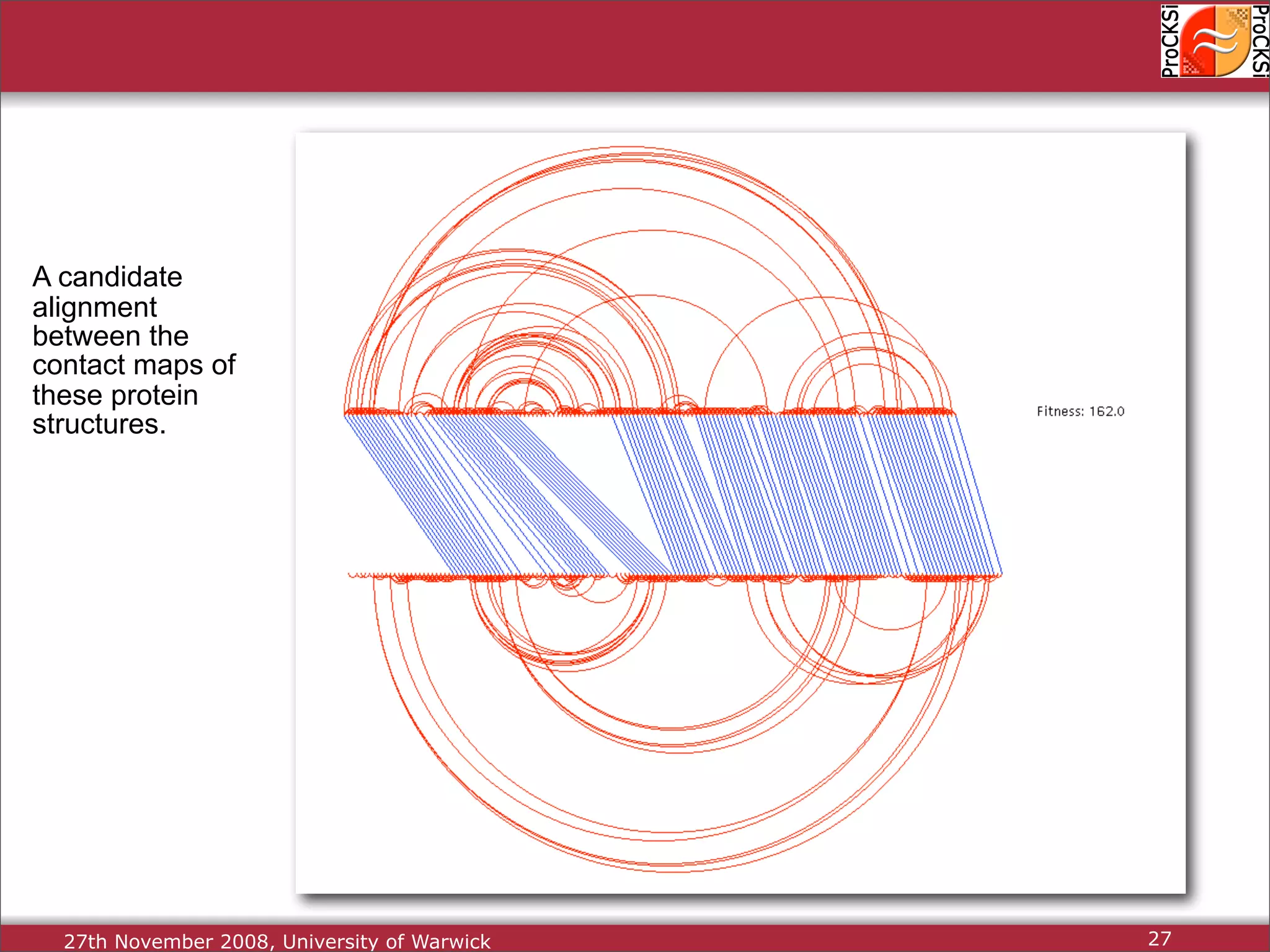
Importance of comparing protein structures, various approaches and algorithms available for comparison.
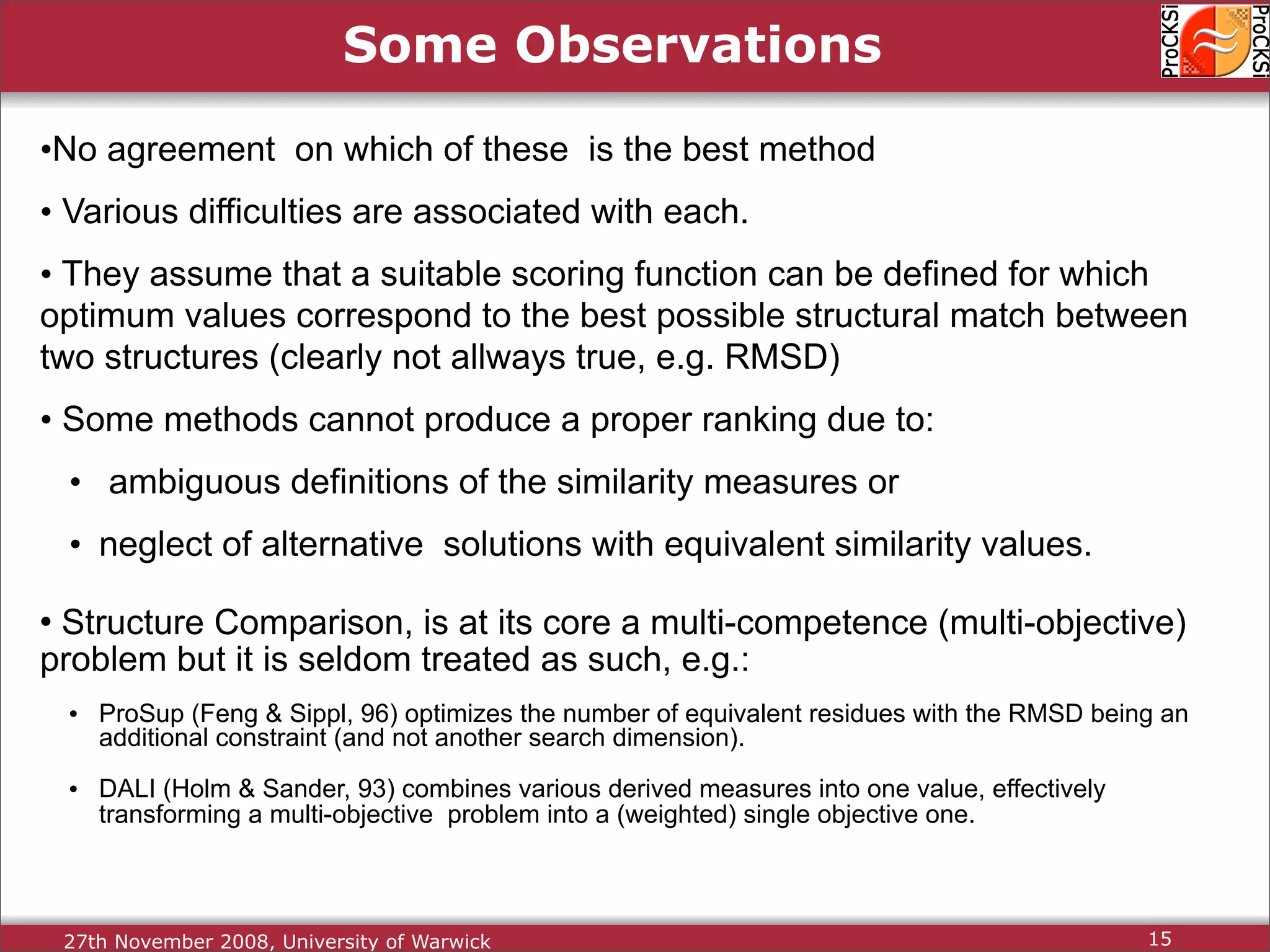
Discussion of dynamic programming and common challenges faced by different protein comparison methods.
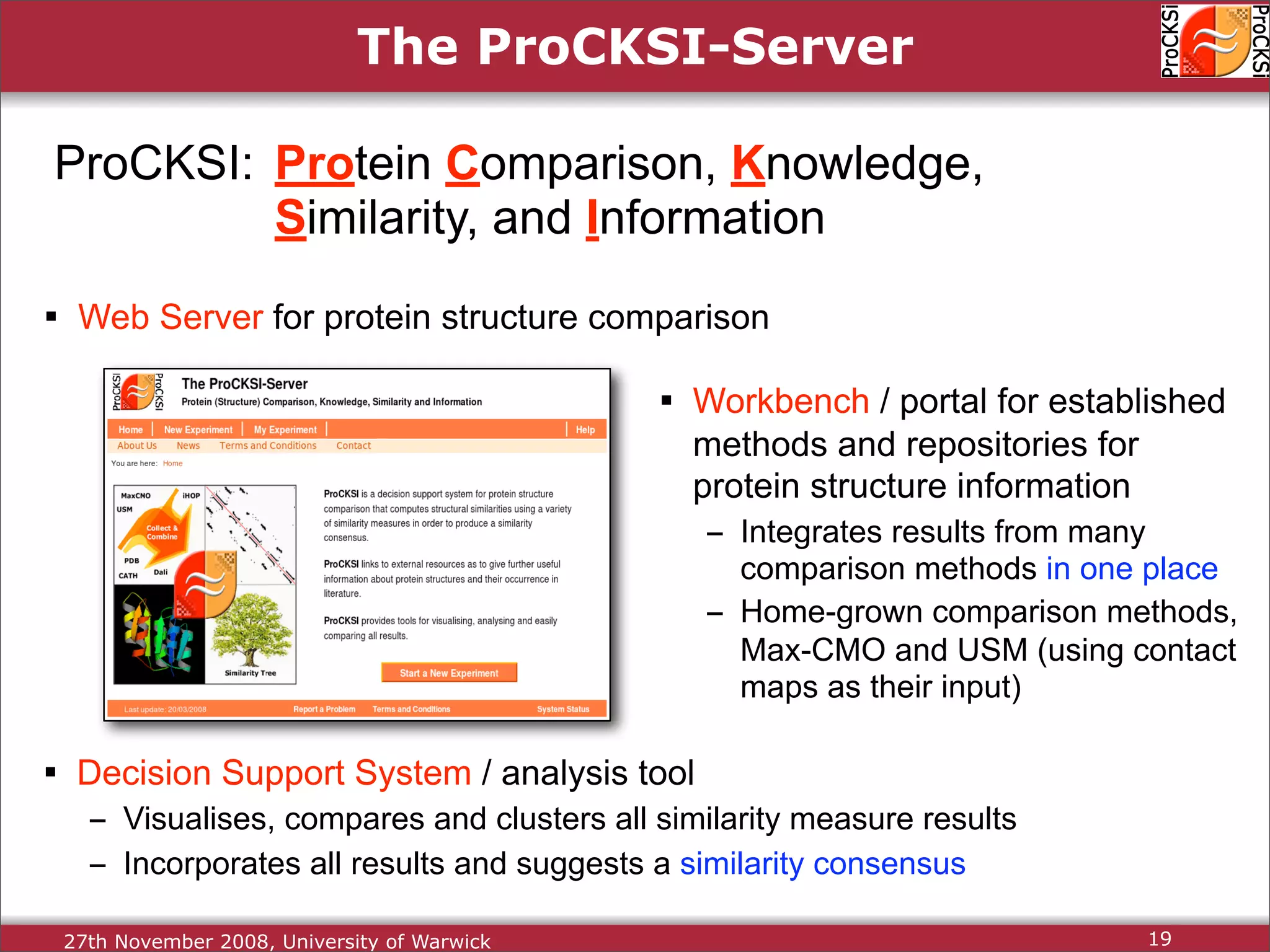
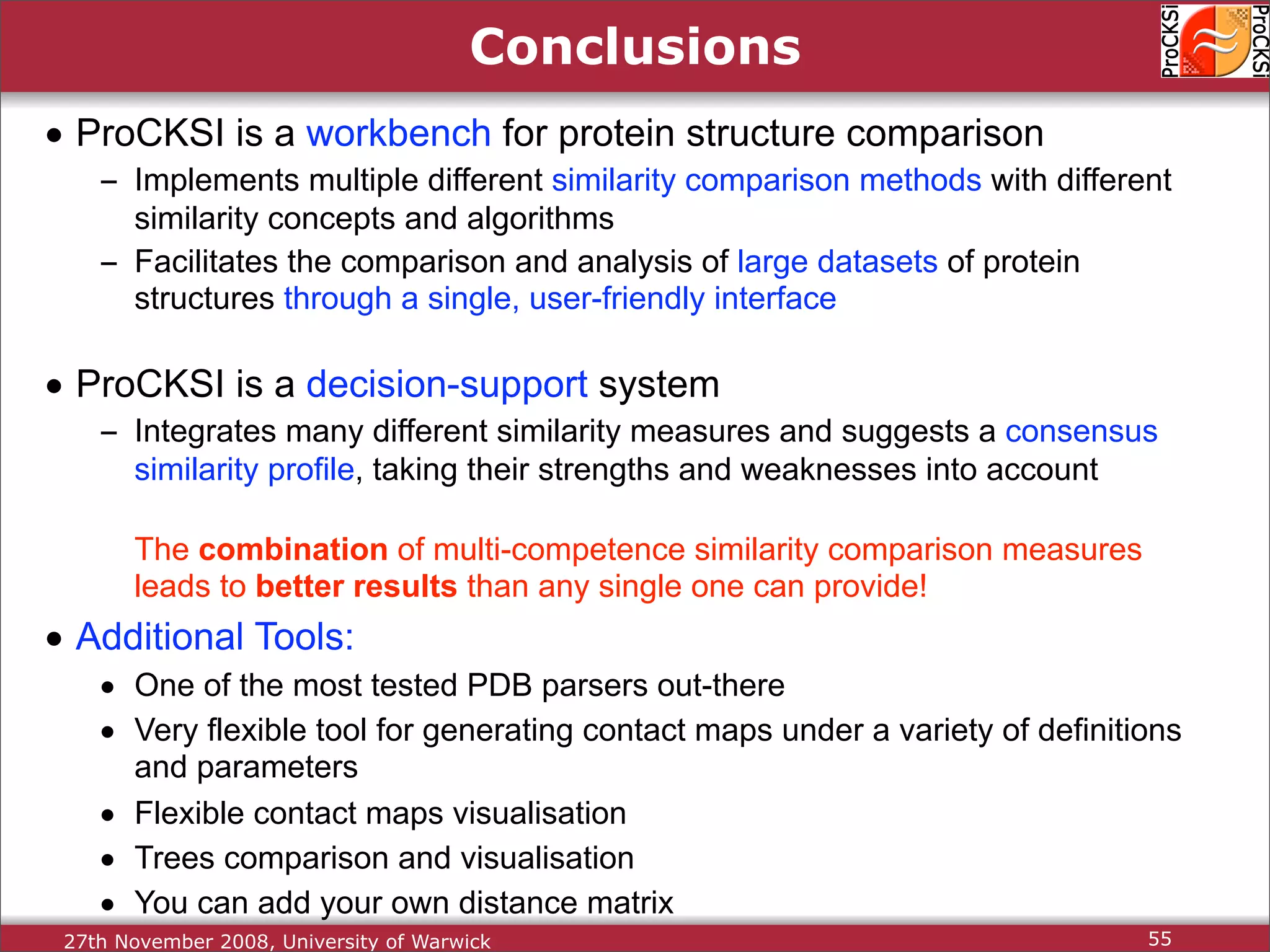
Introduction to ProCKSI as a comprehensive web server for protein comparison and integration of methods.
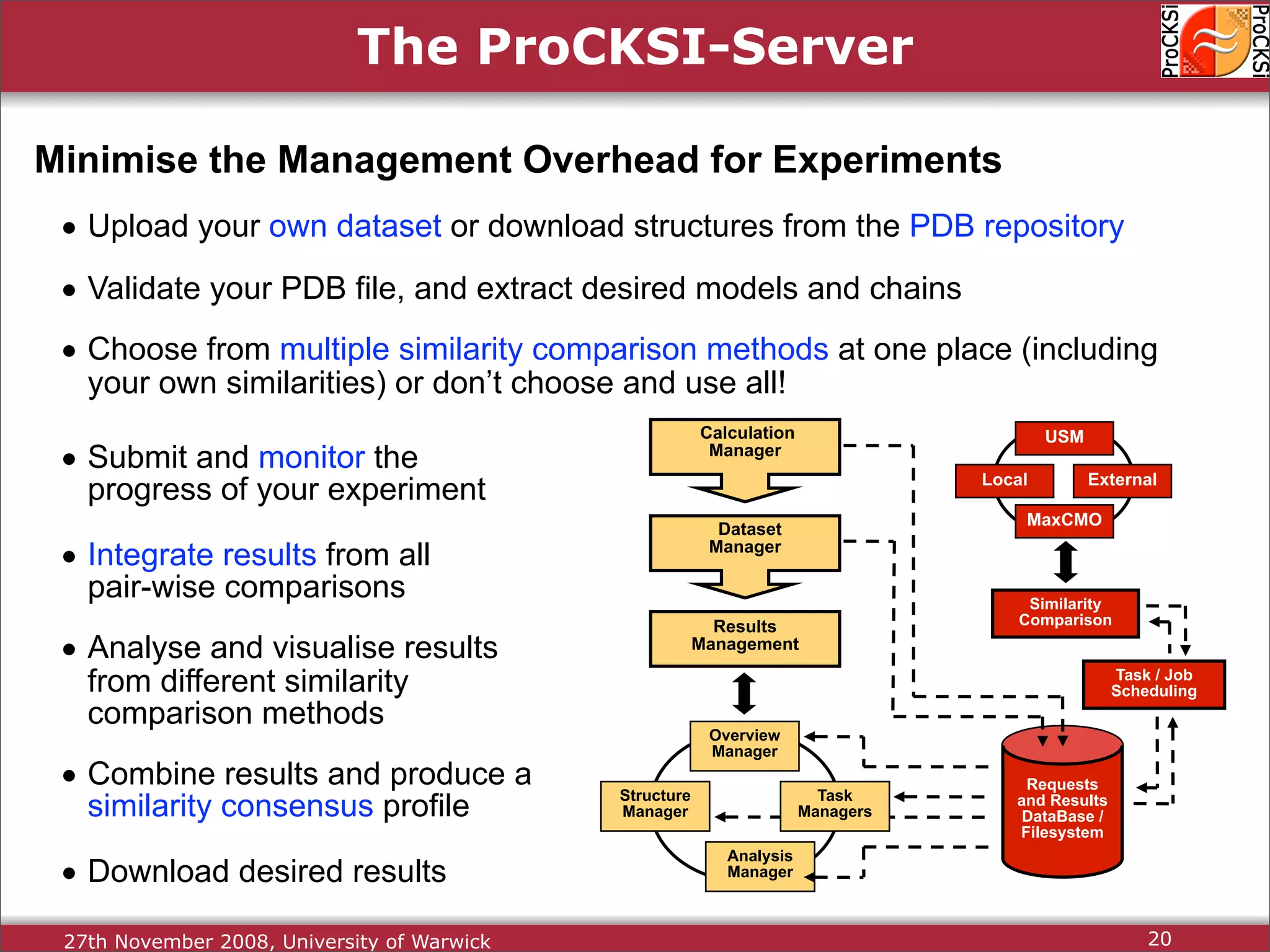
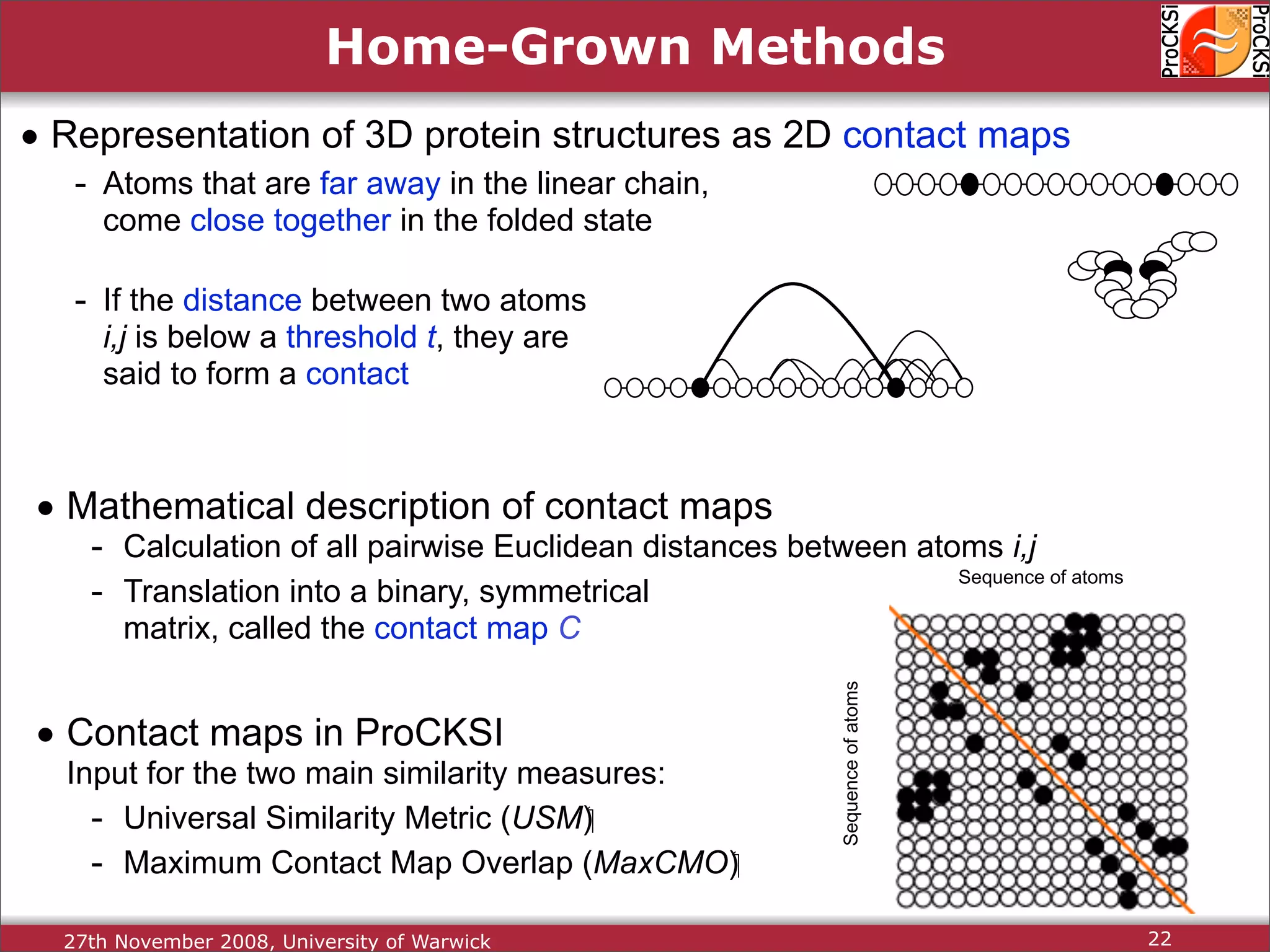
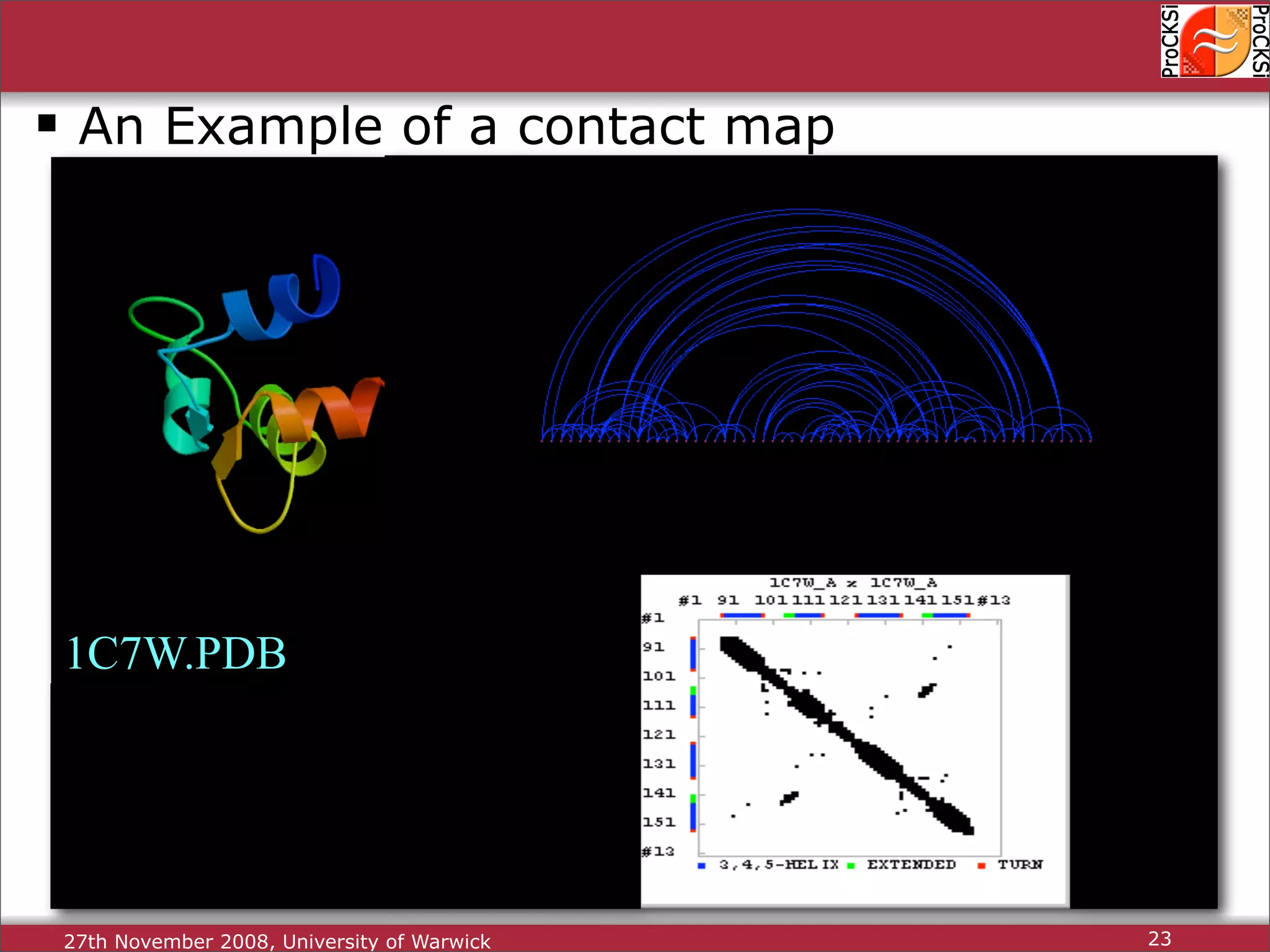
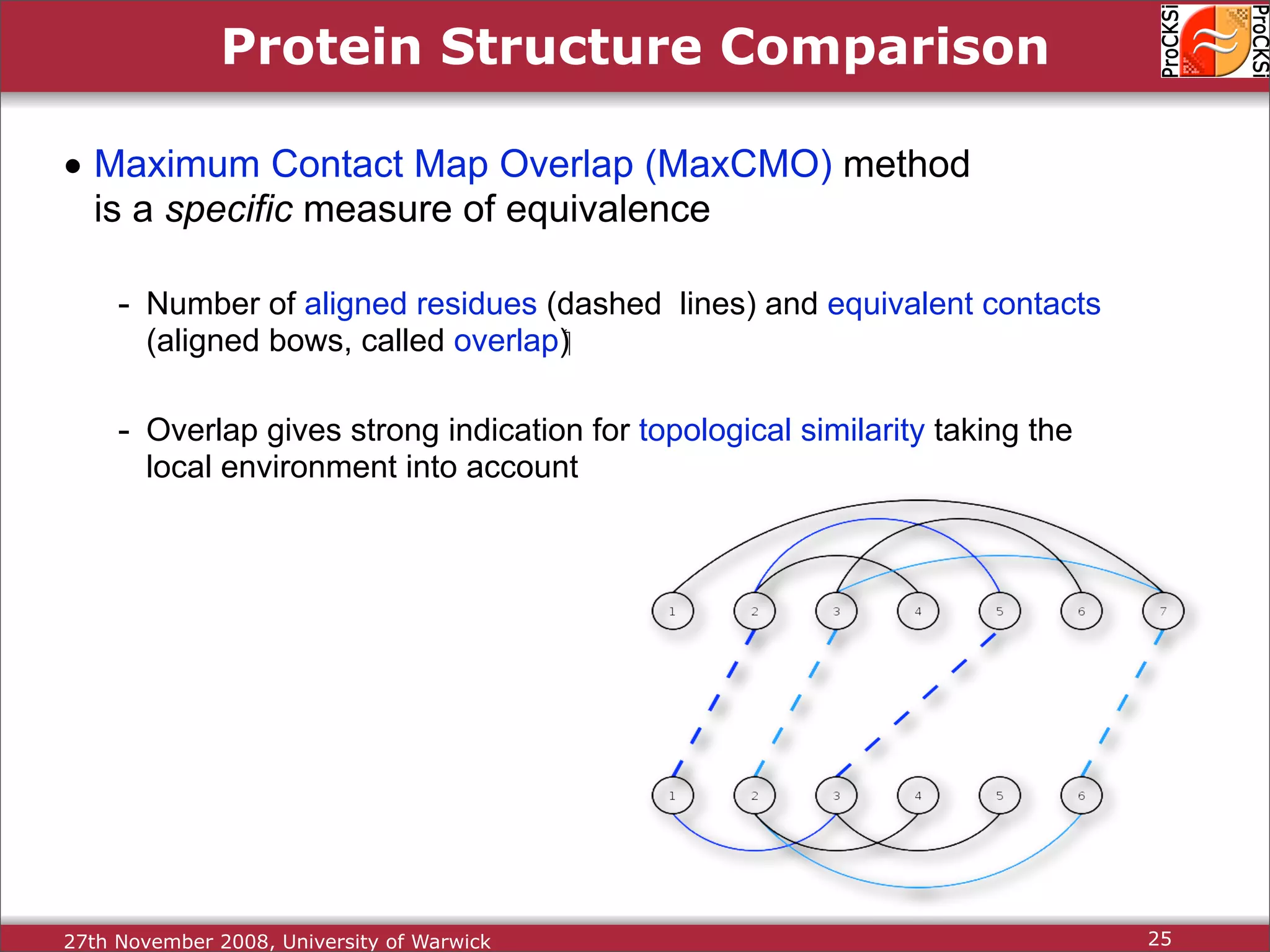
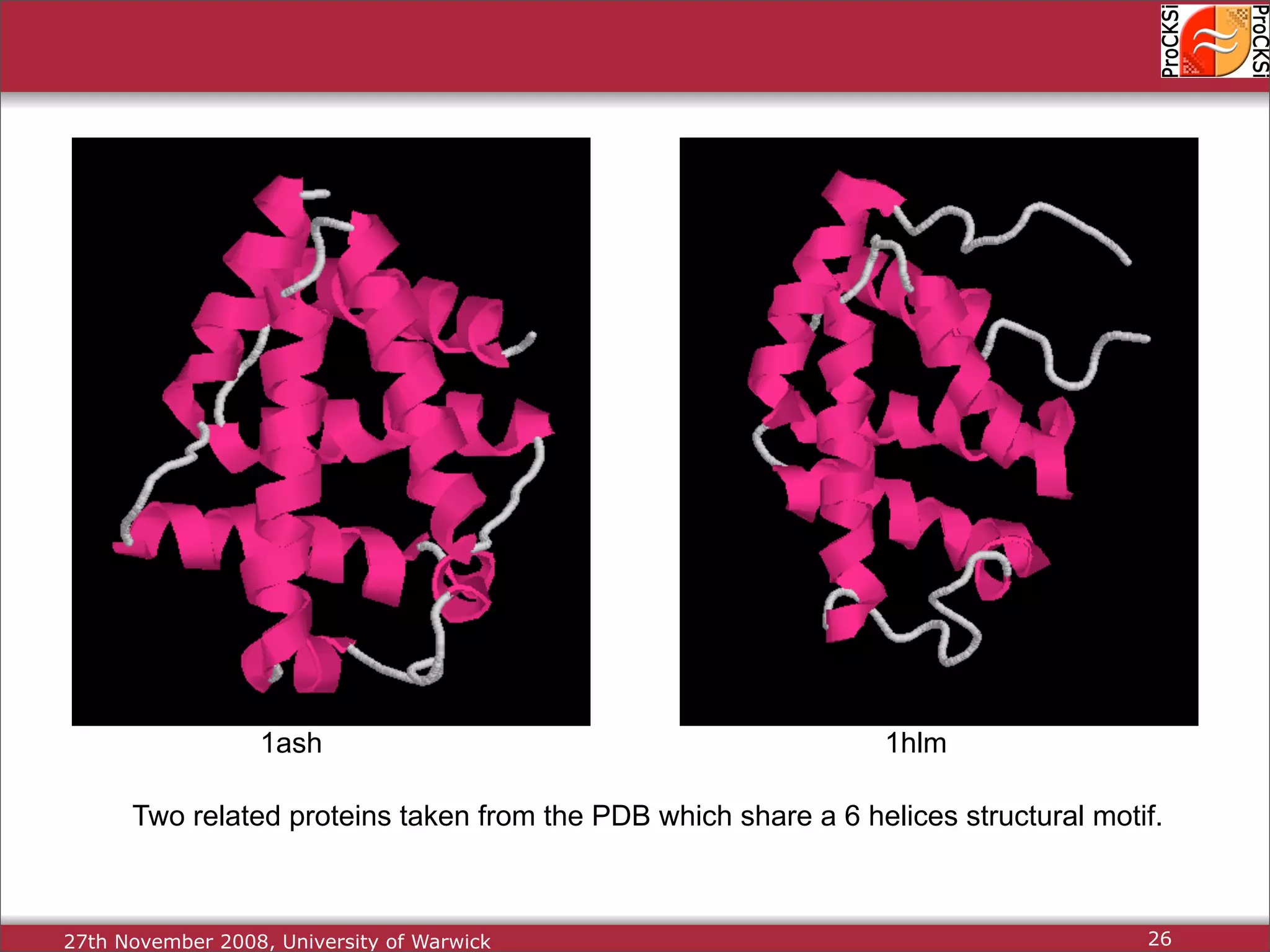
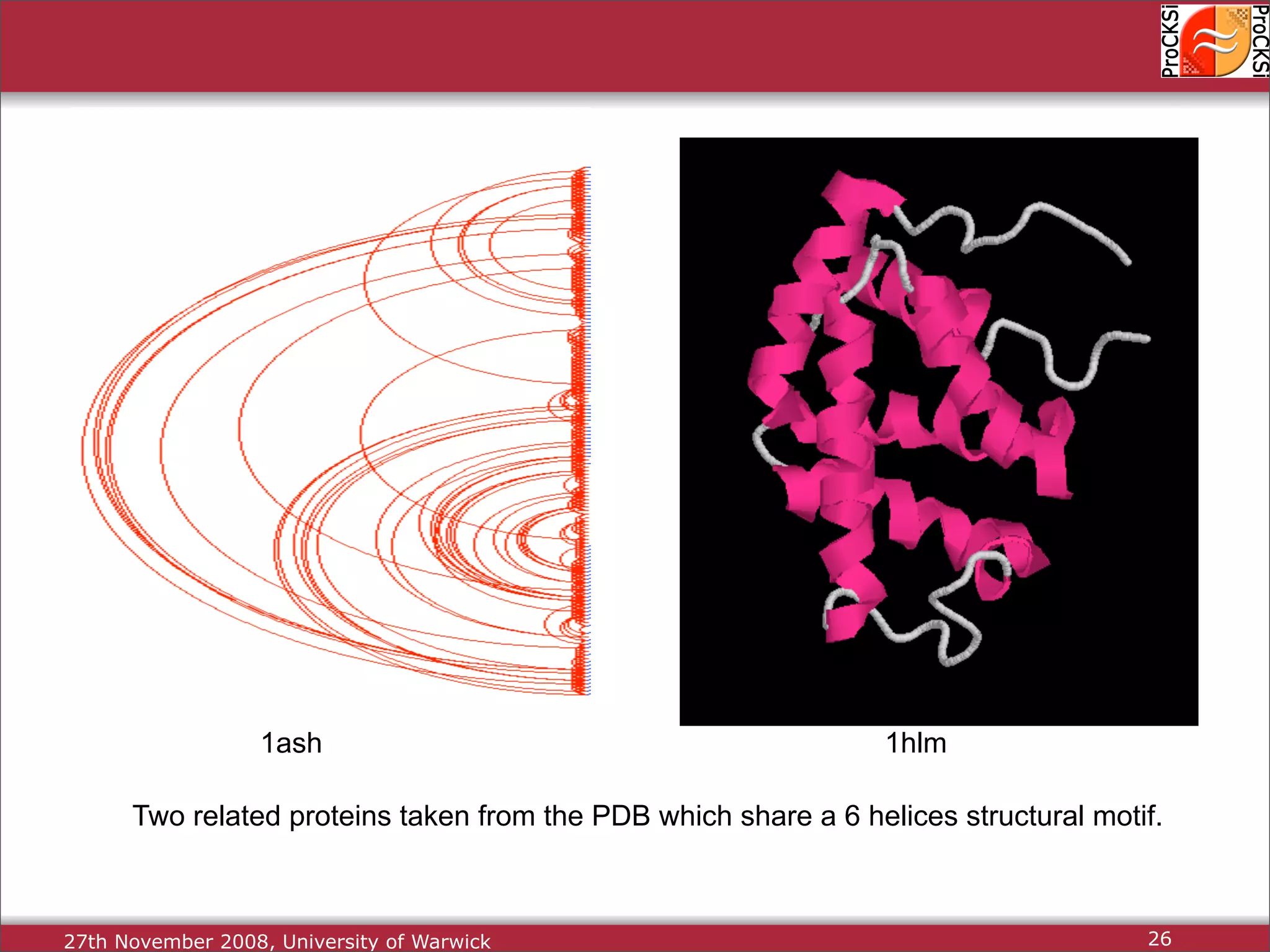
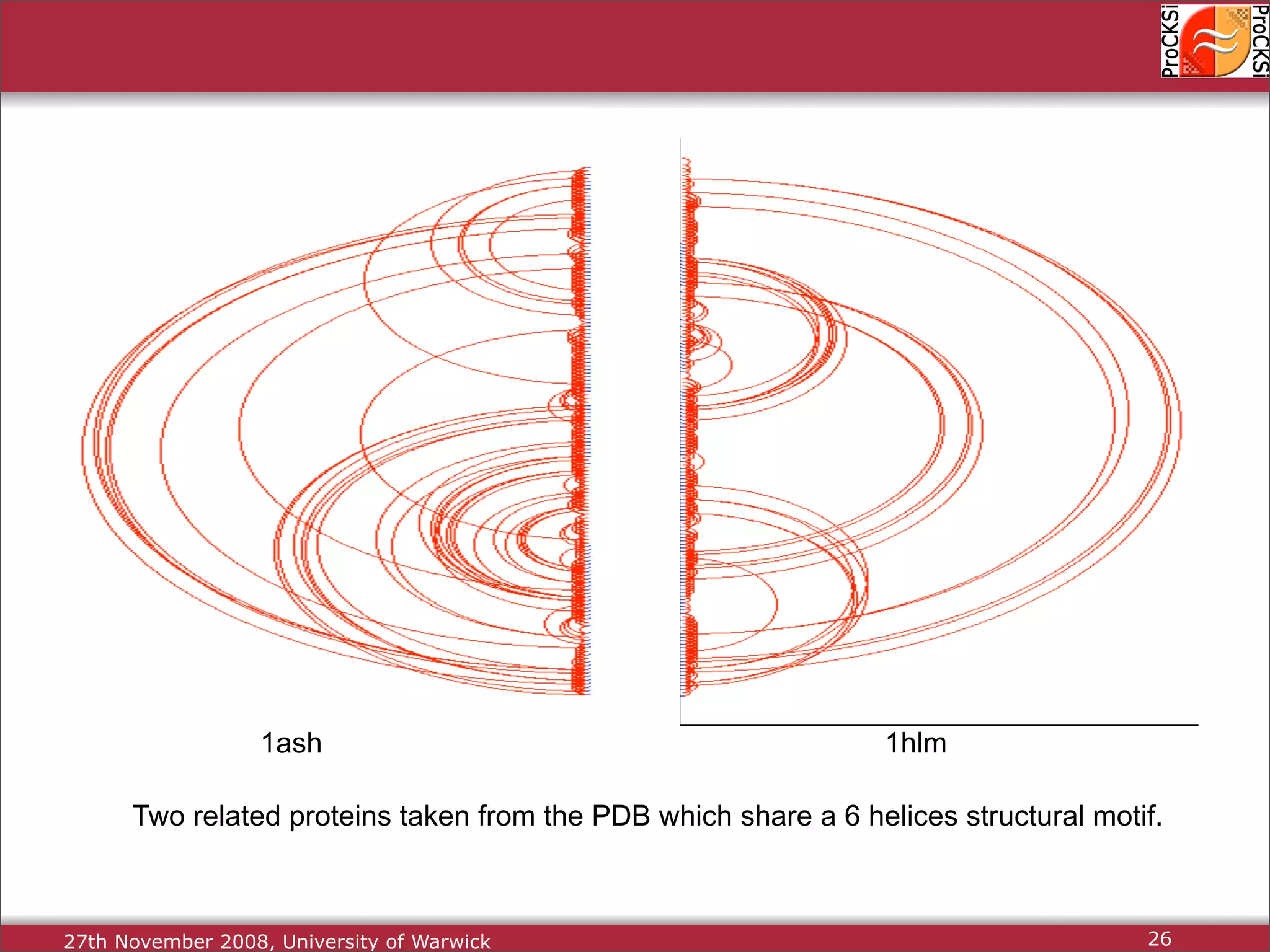
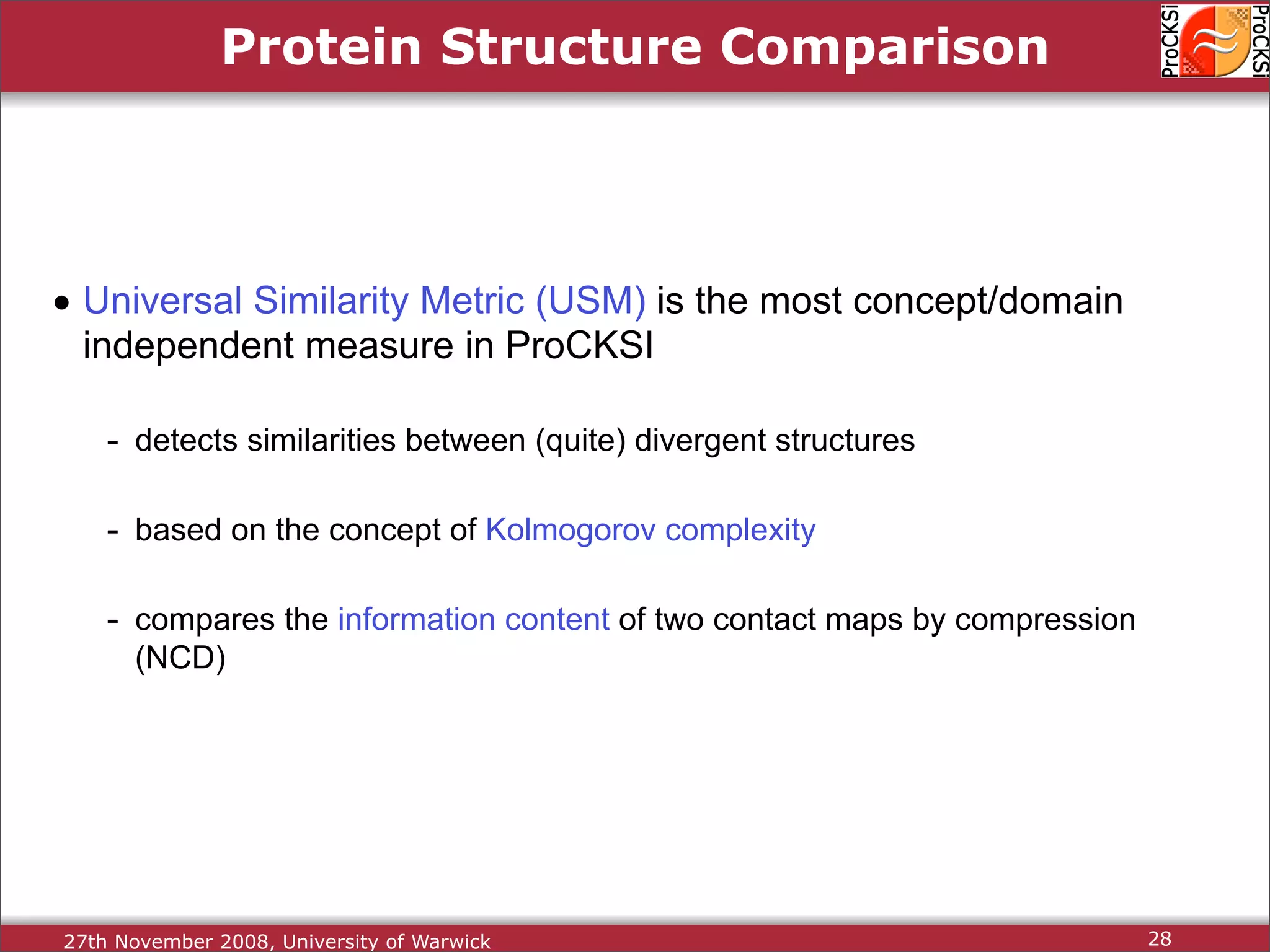
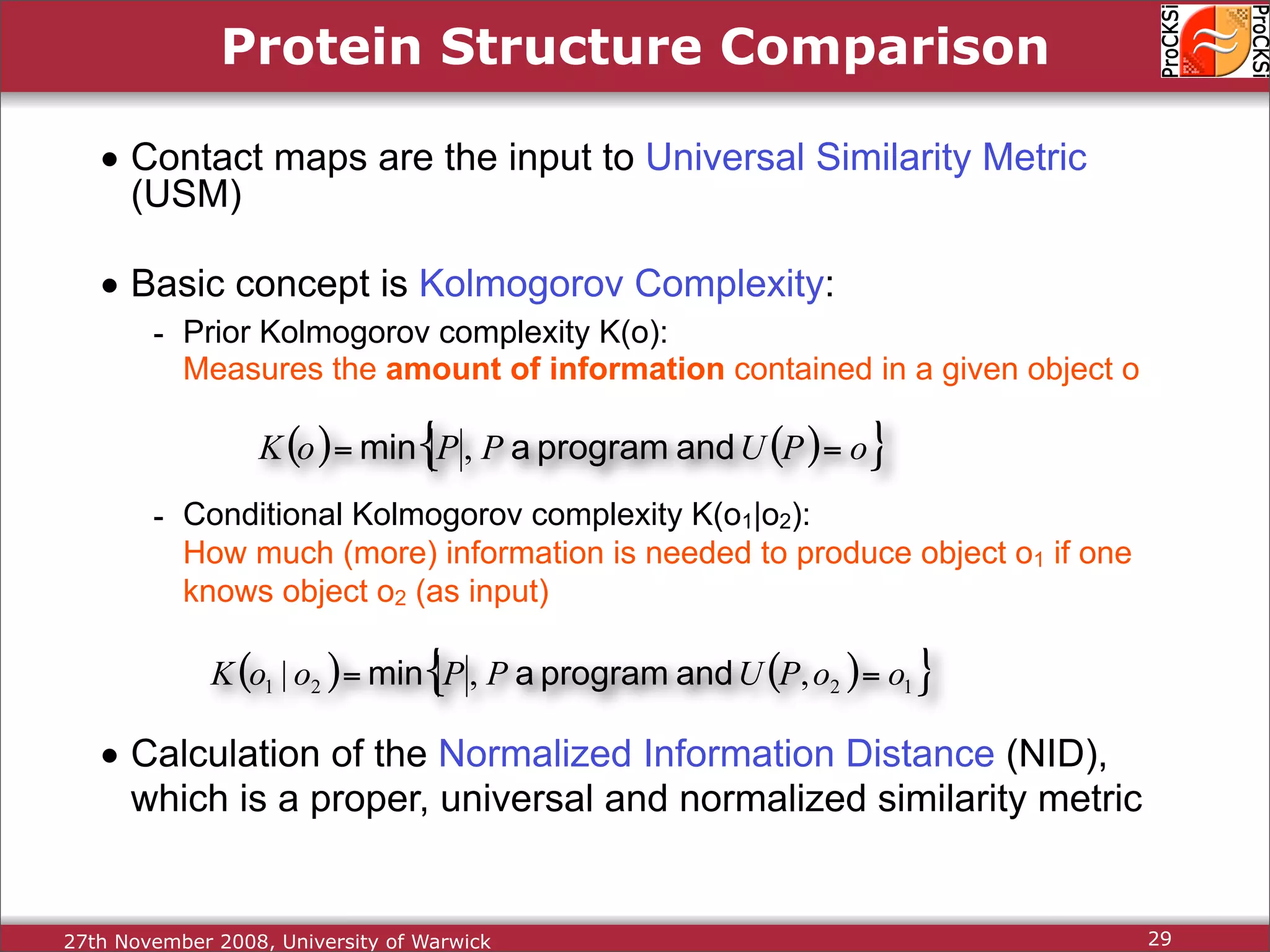
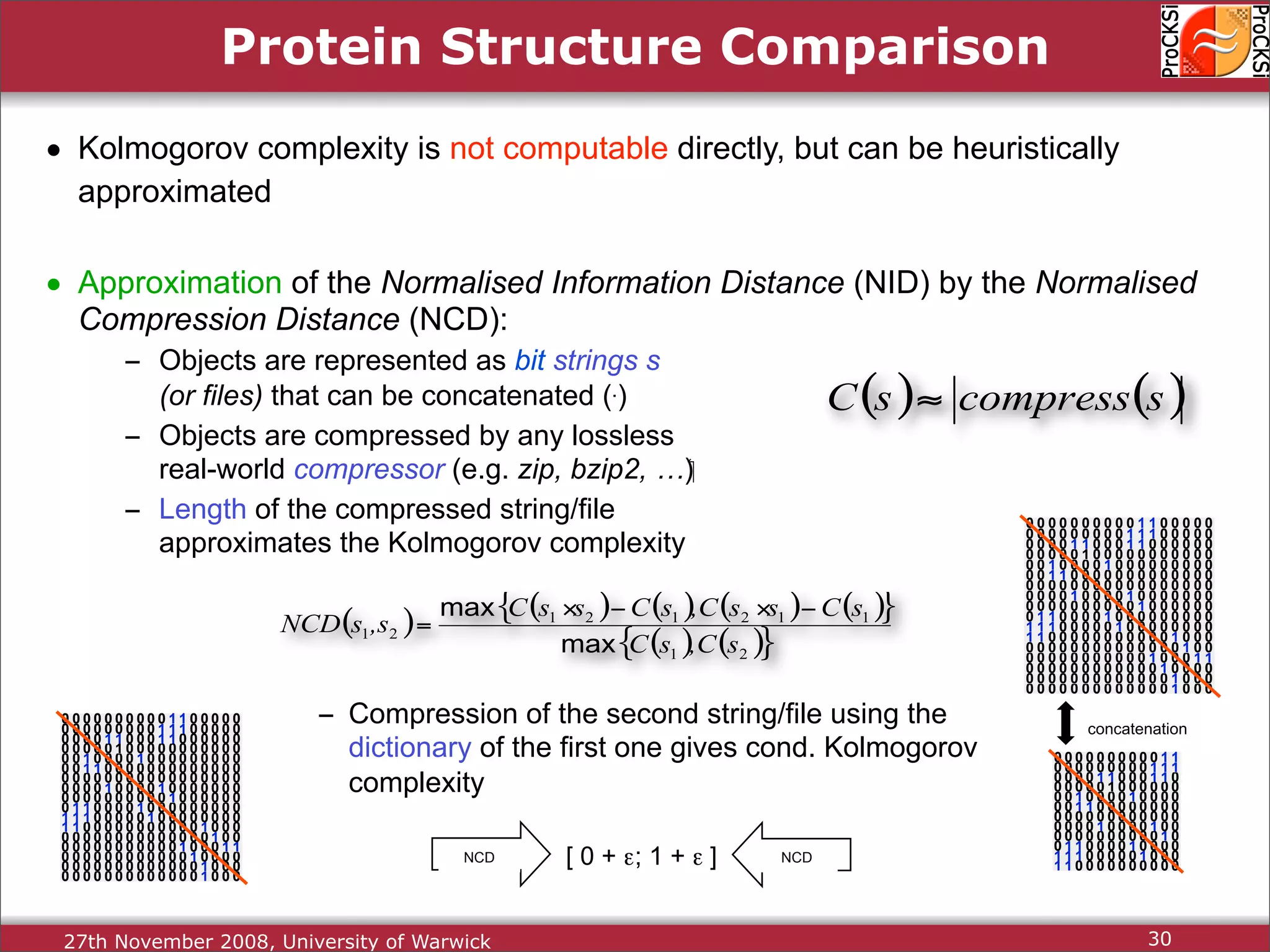
Detailed insights into home-grown methods (USM, Max-CMO) and process of validating and extracting data.
Use of universal similarity metric (USM) and methods of analyzing protein structure comparisons.
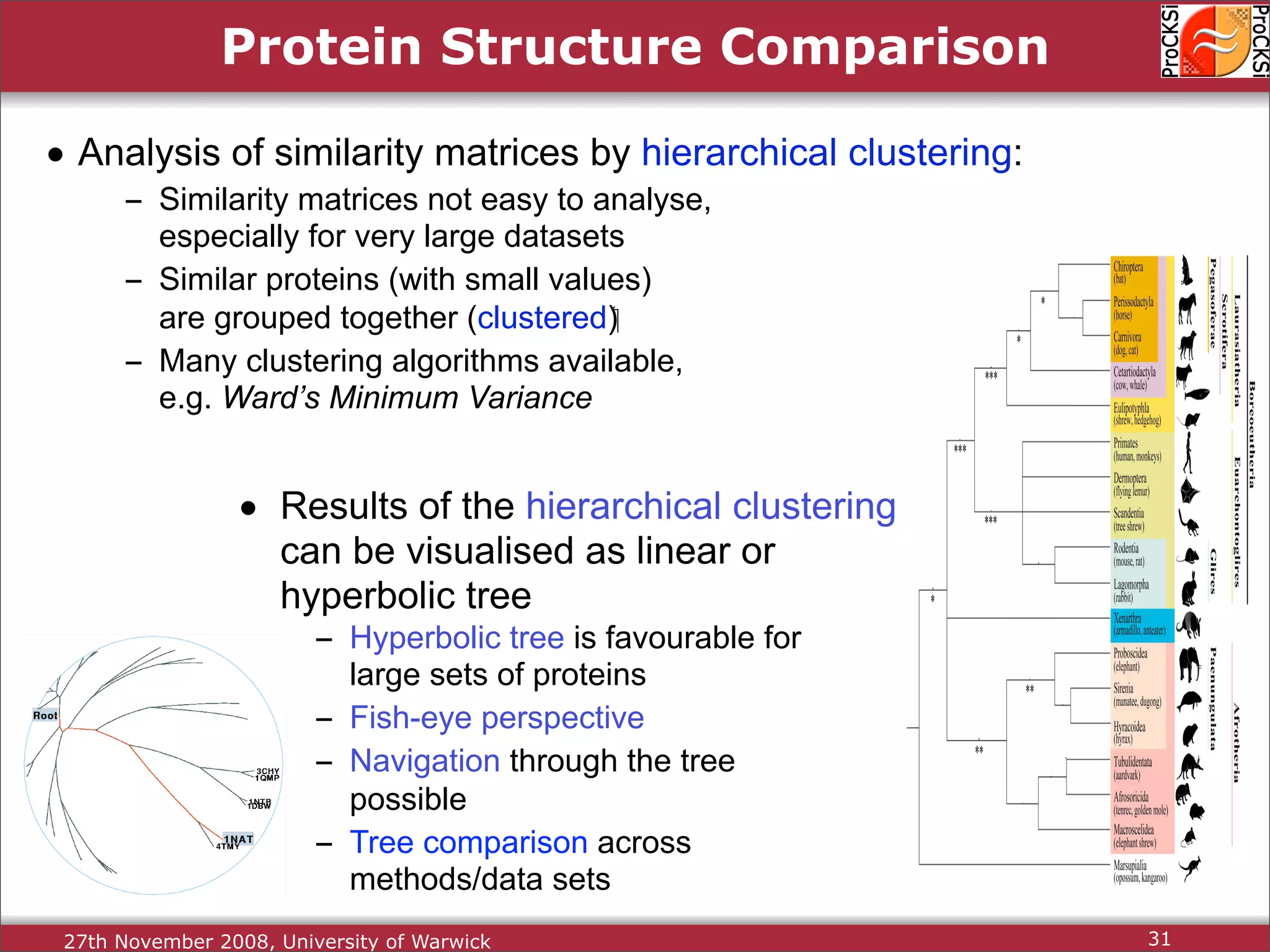
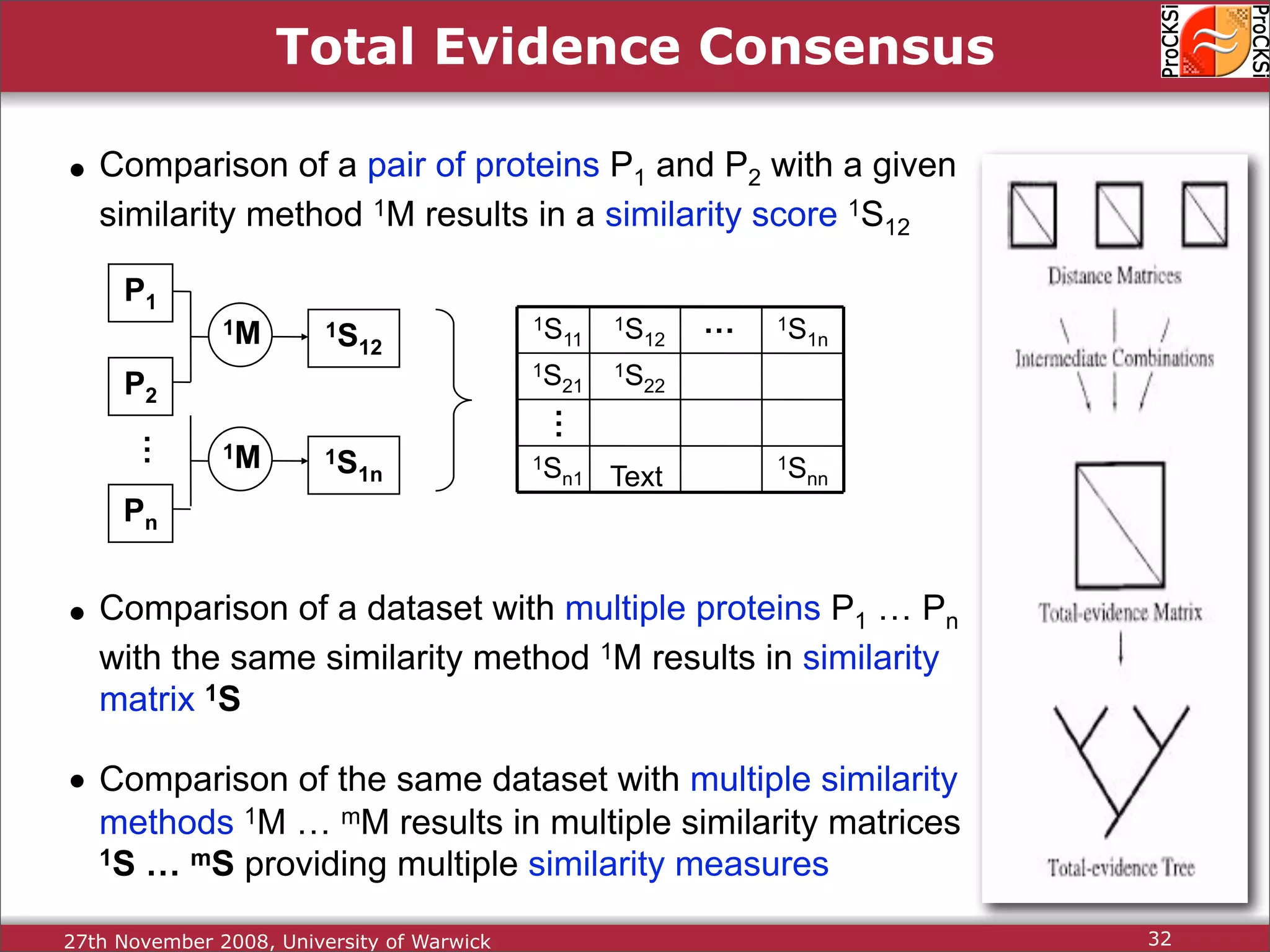
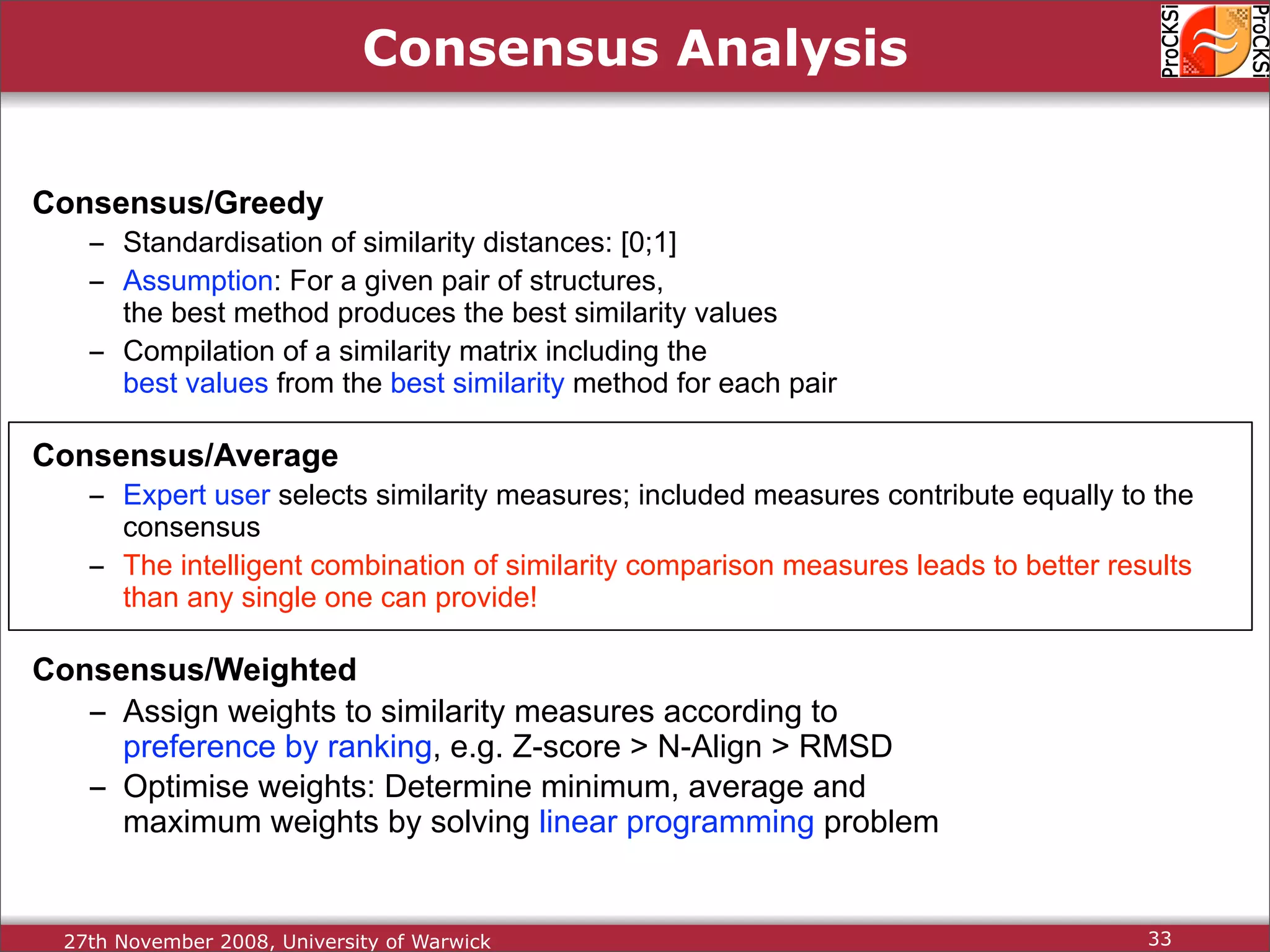
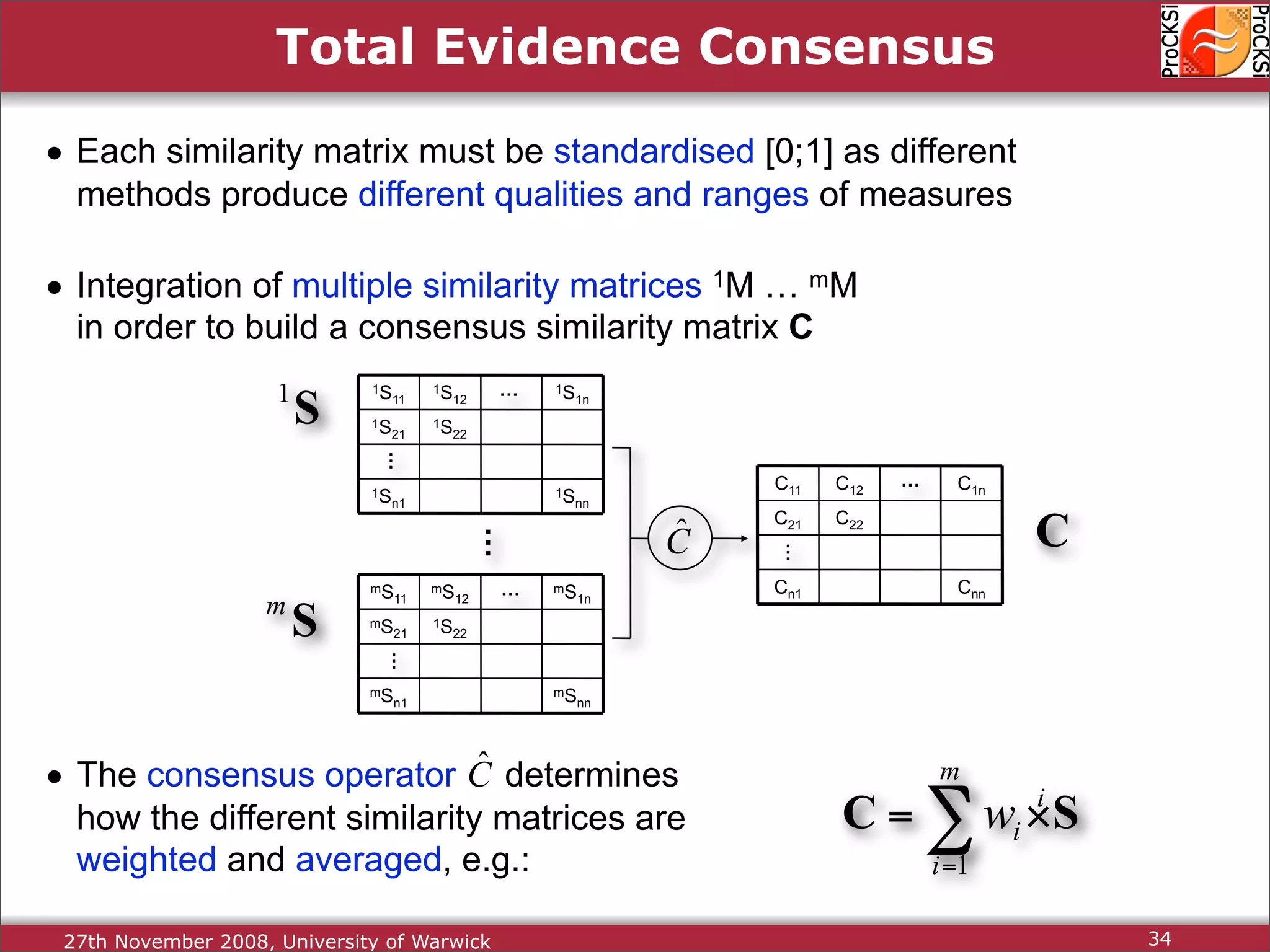
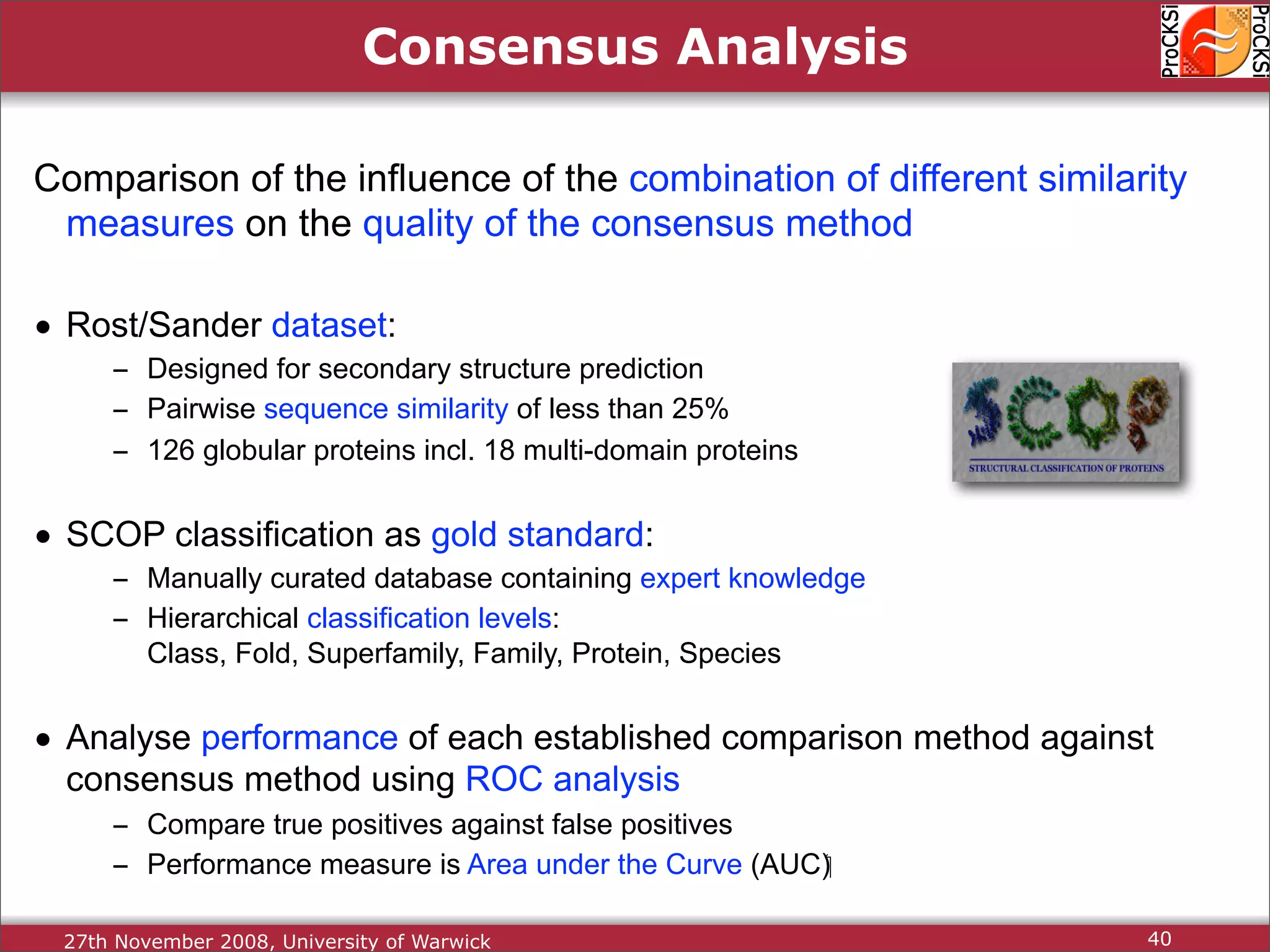
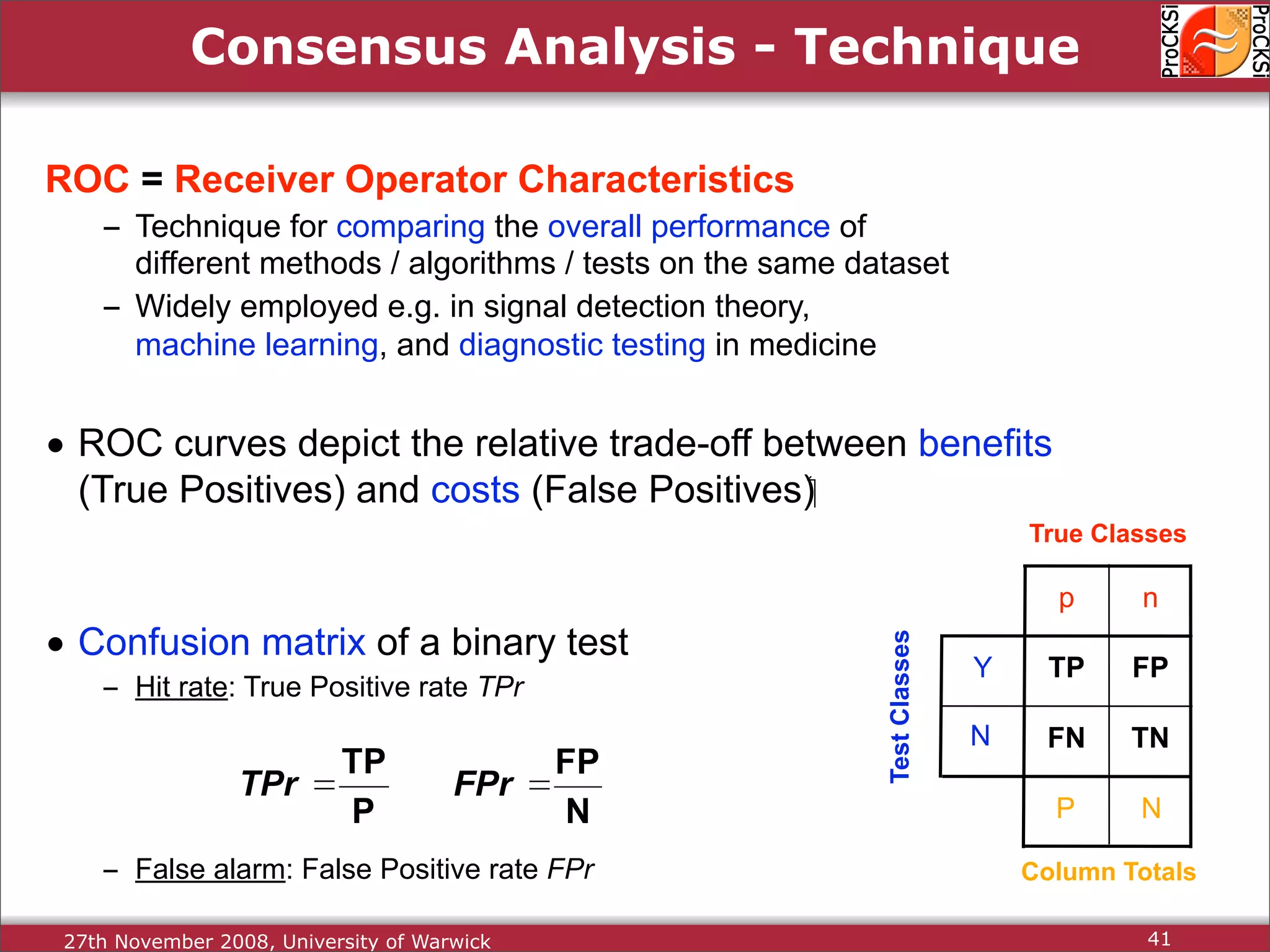
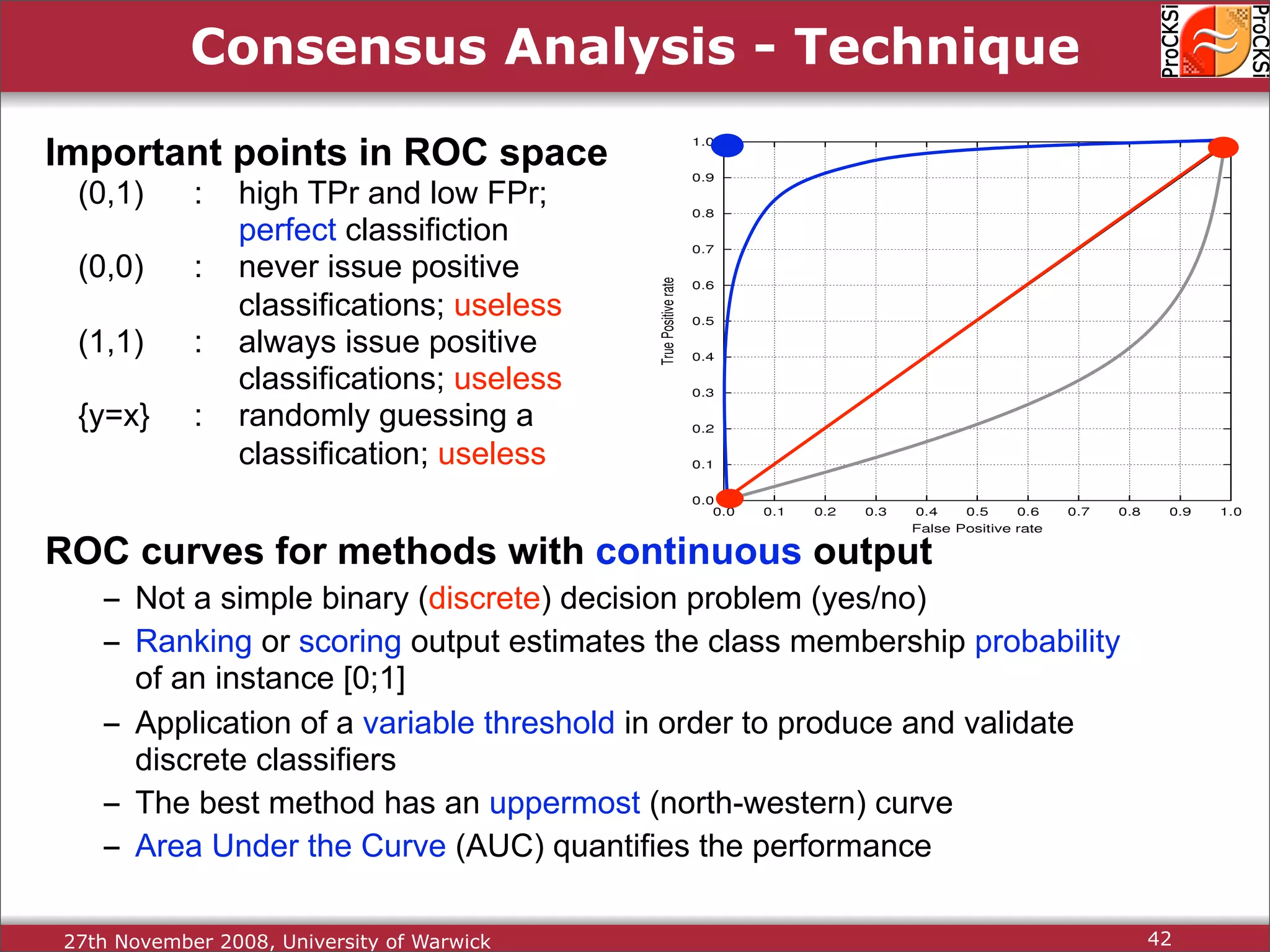
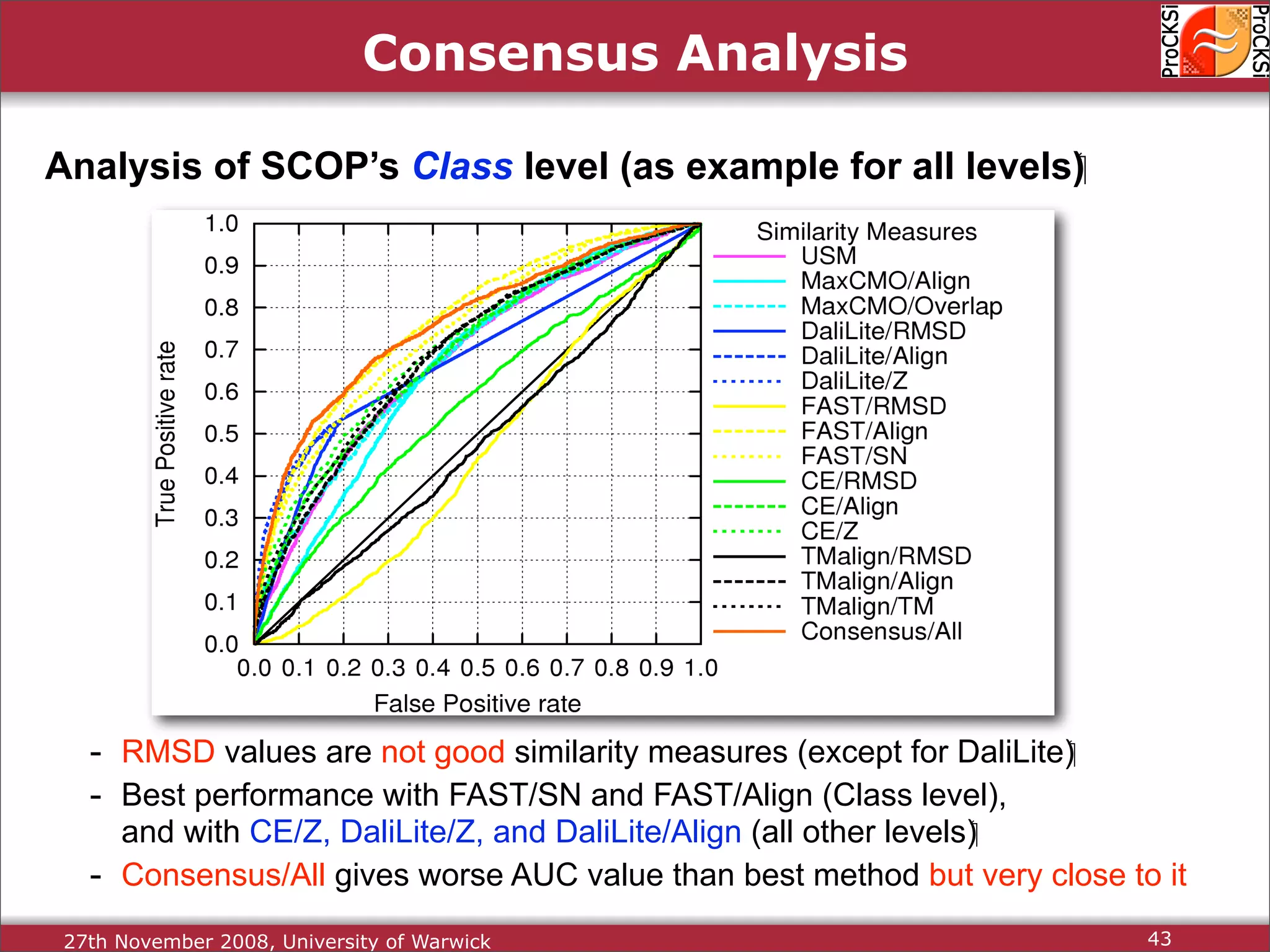
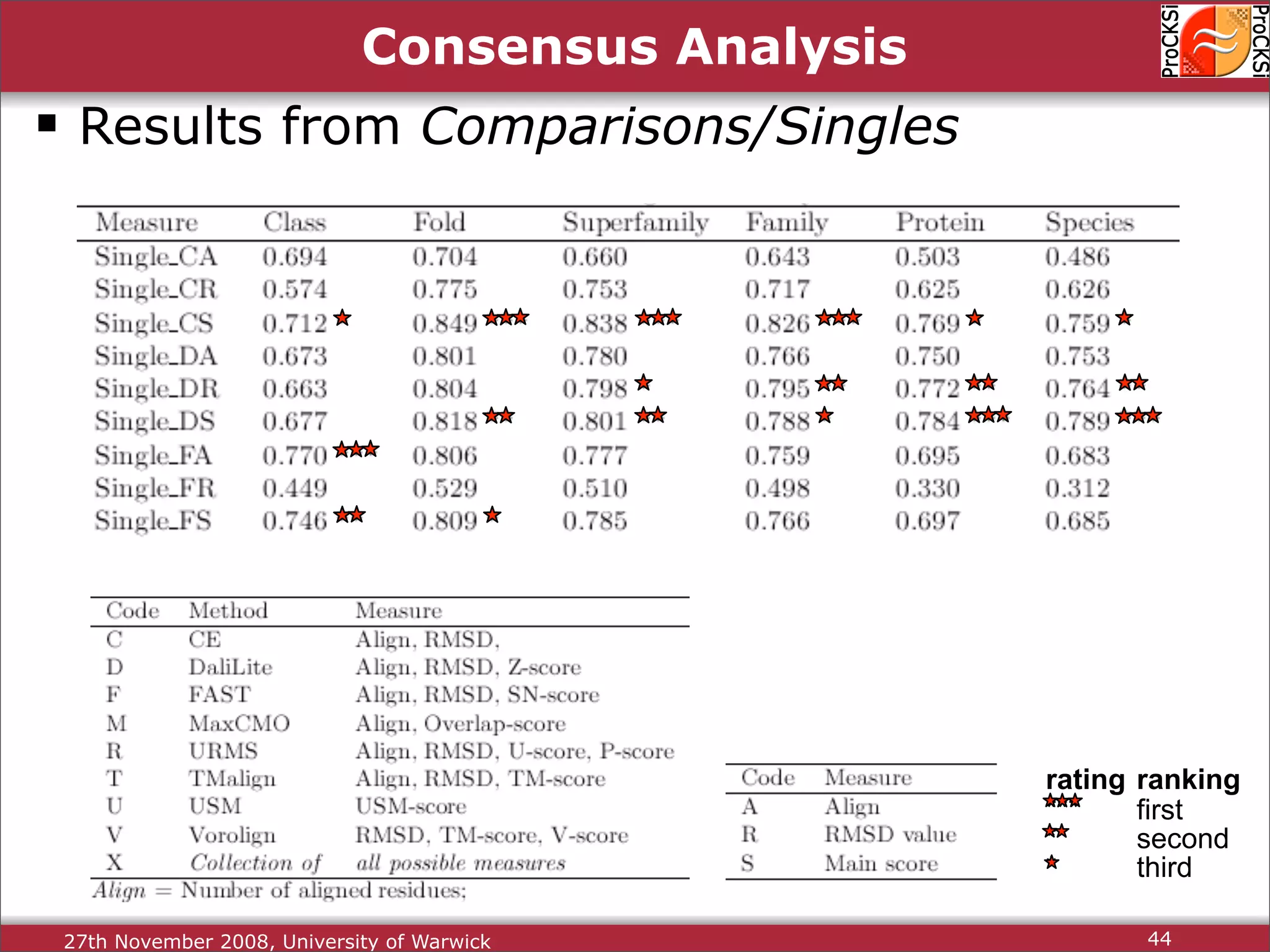
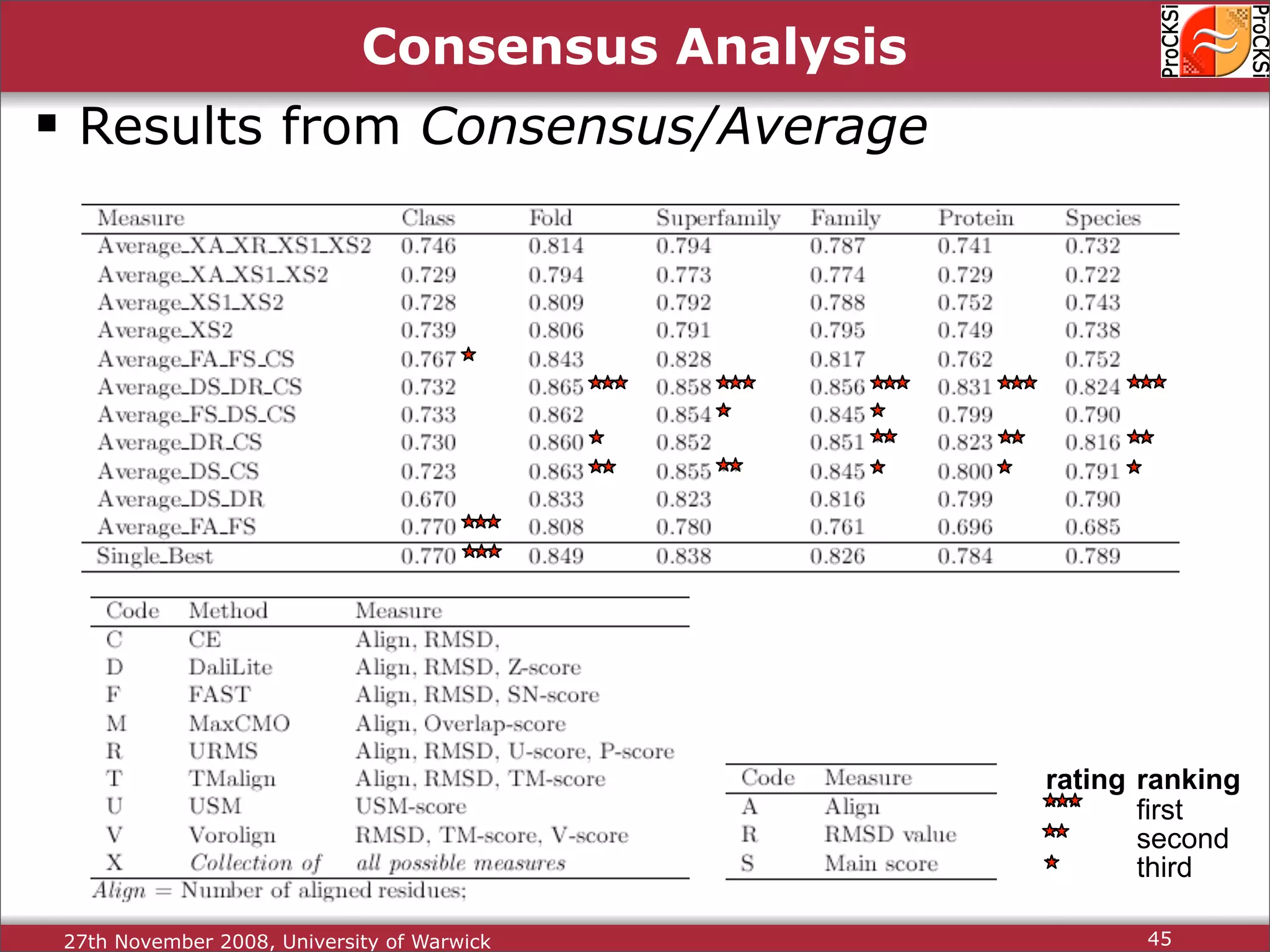
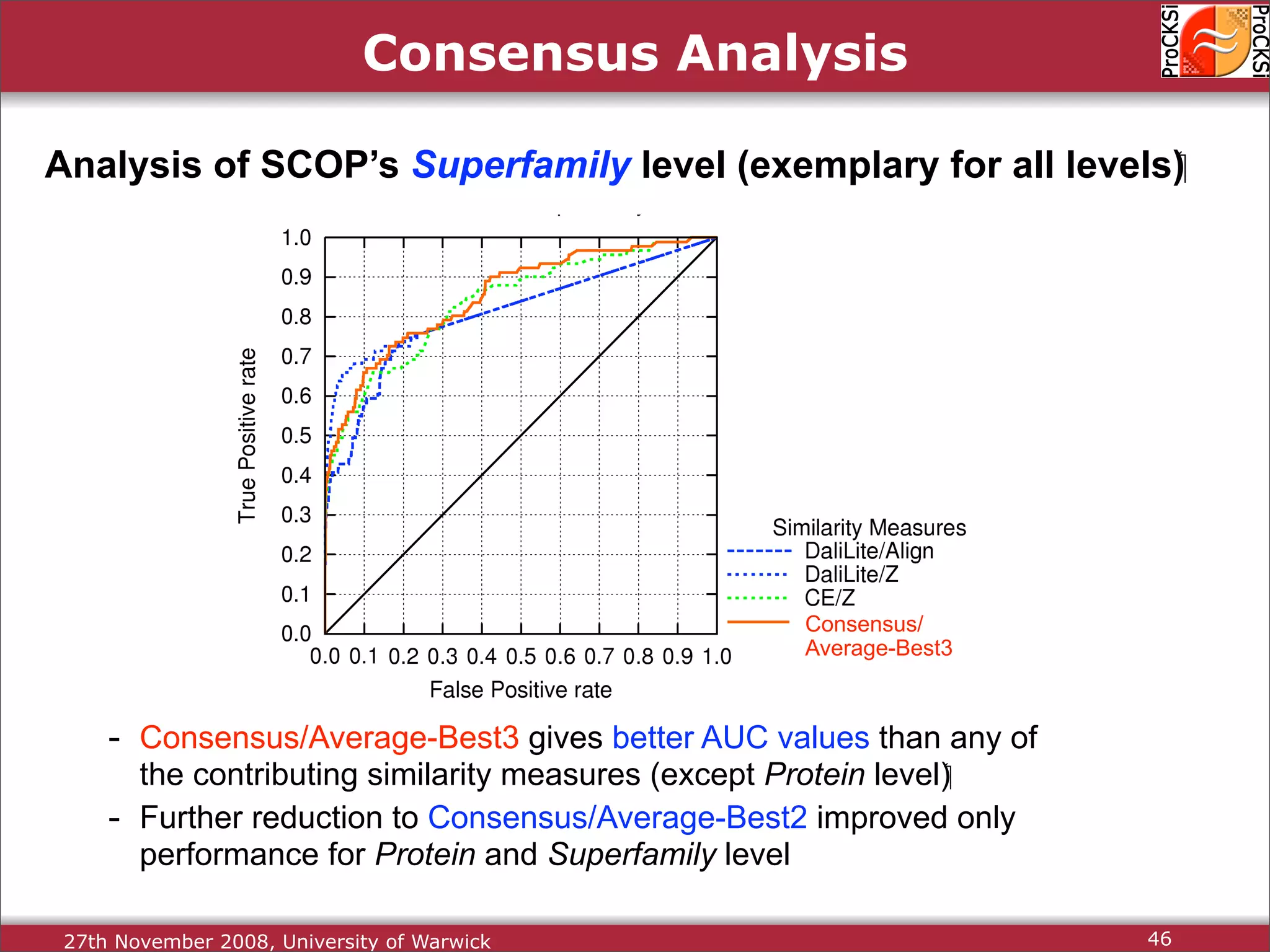
Discussion on multiple consensus analysis approaches in integrating similarity matrices for better insights.
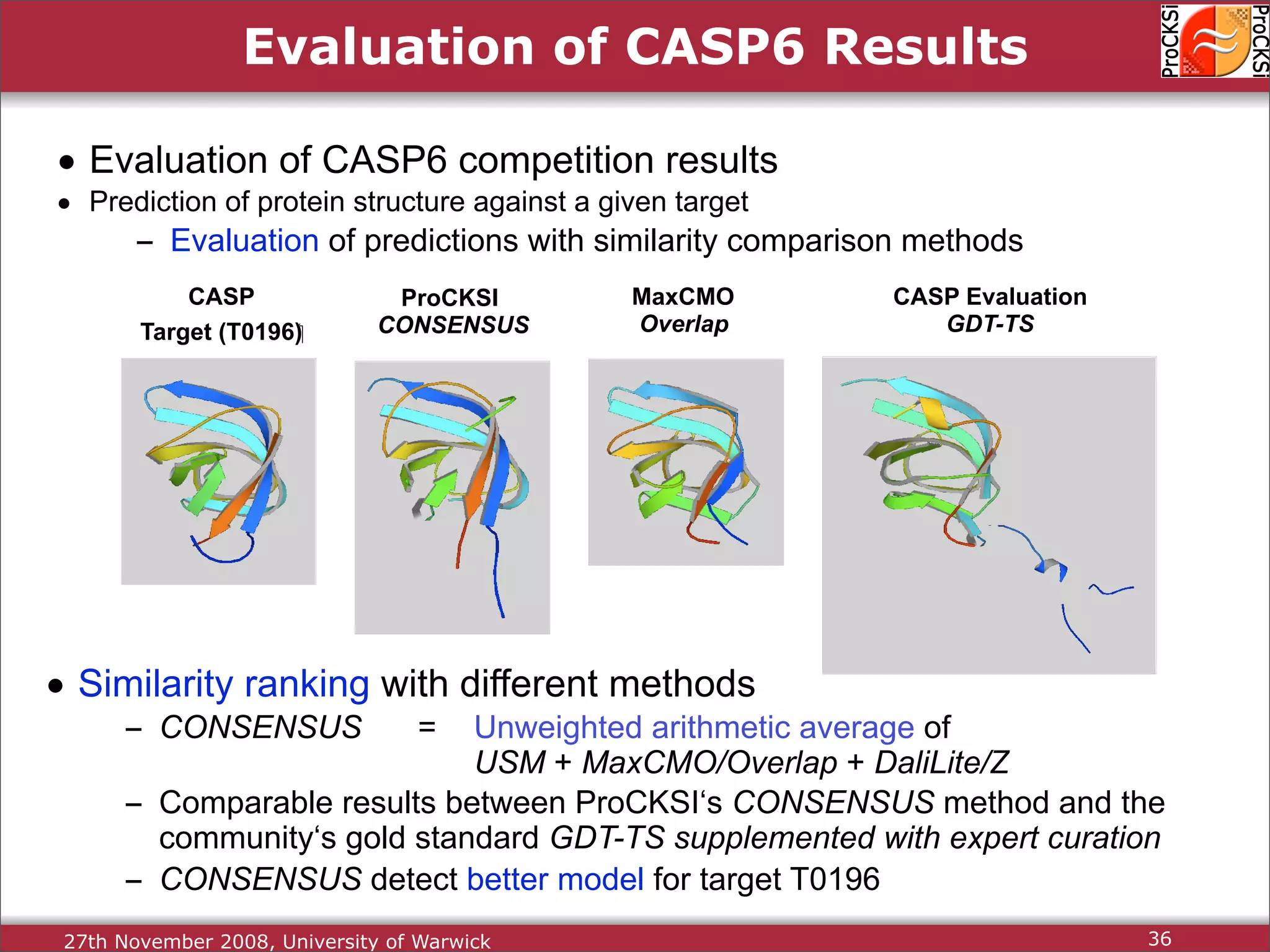
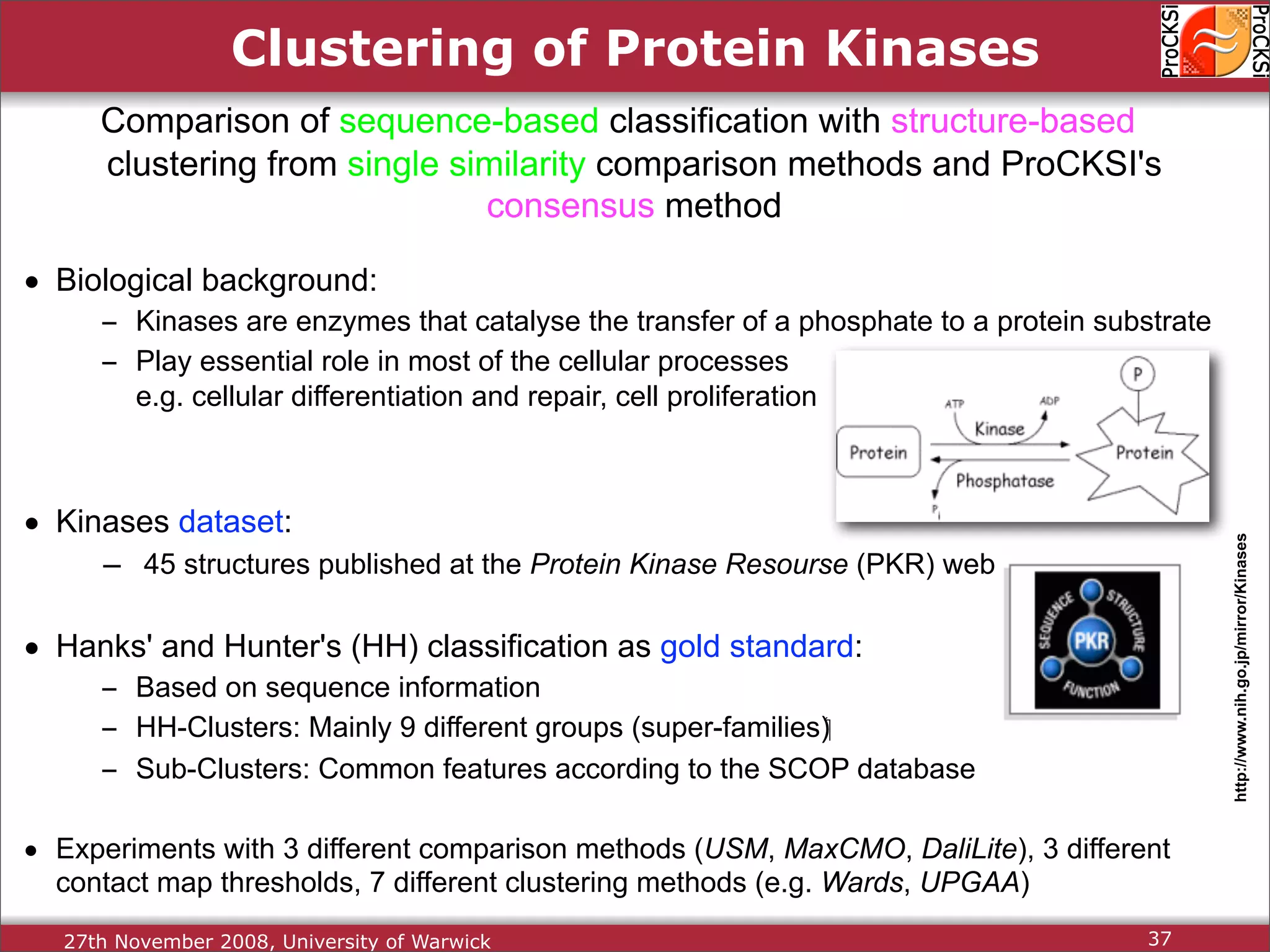
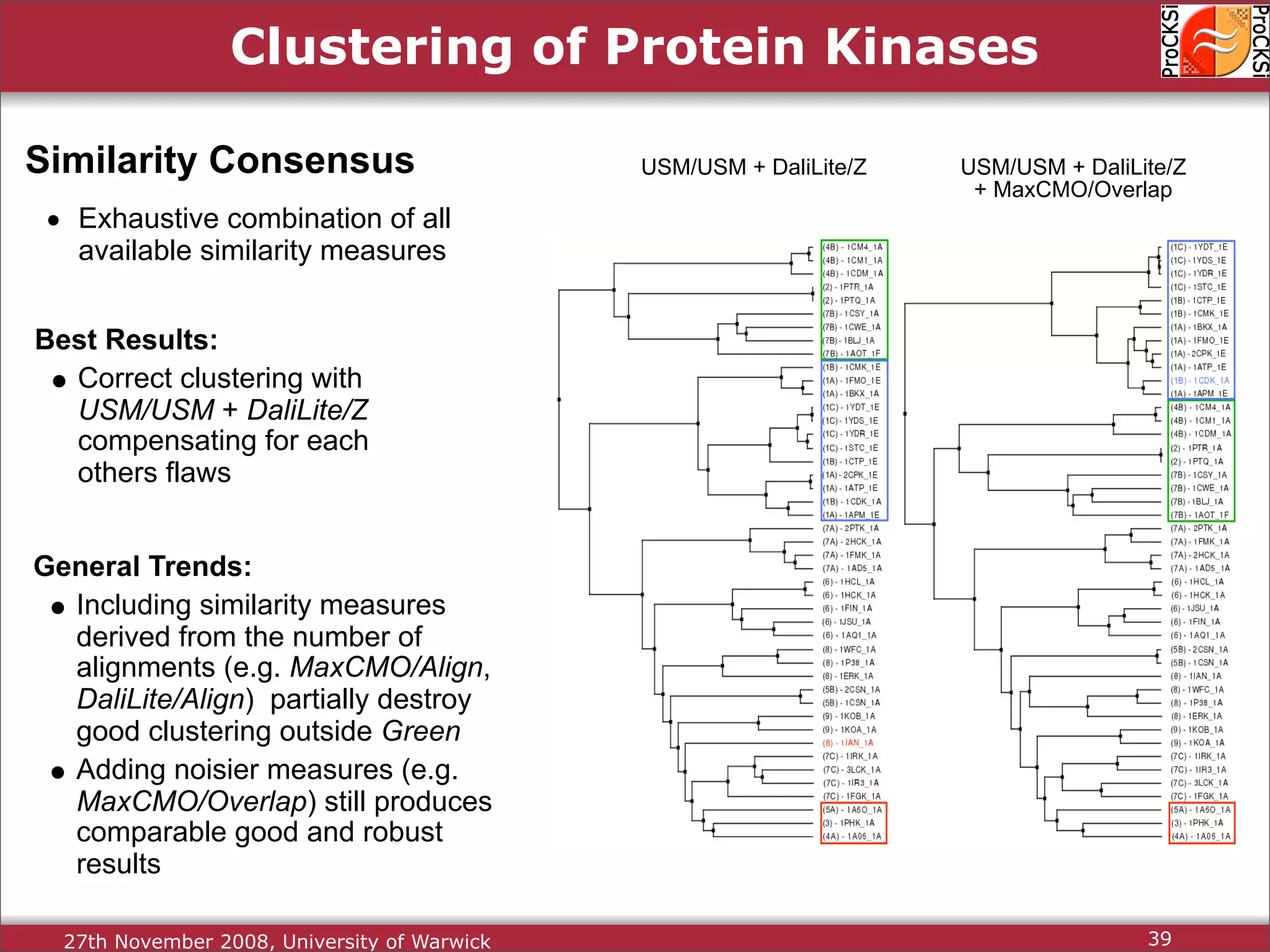
Evaluation of results from CASP6 and clustering of kinases using different comparison methods, assessing accuracy.
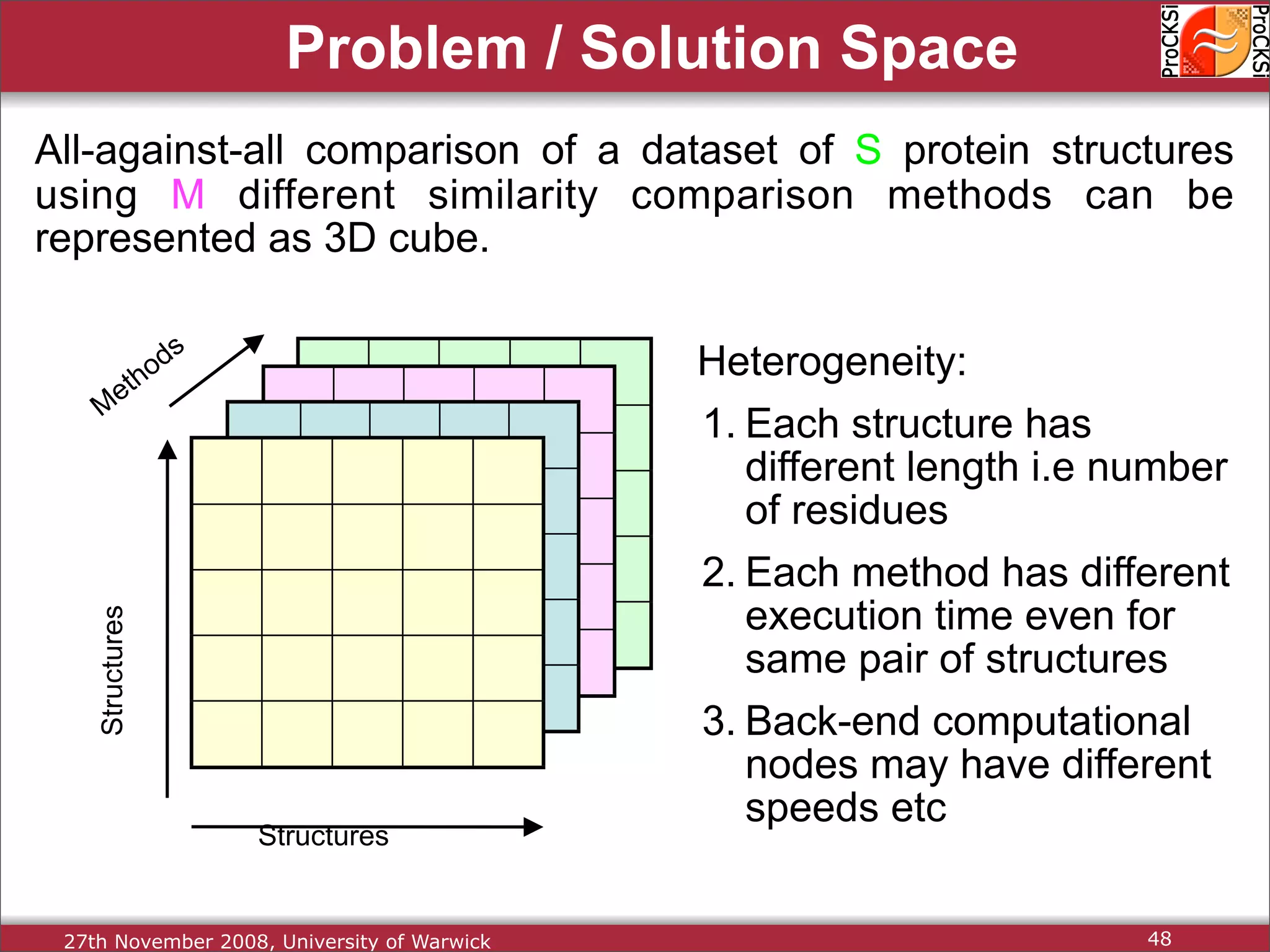
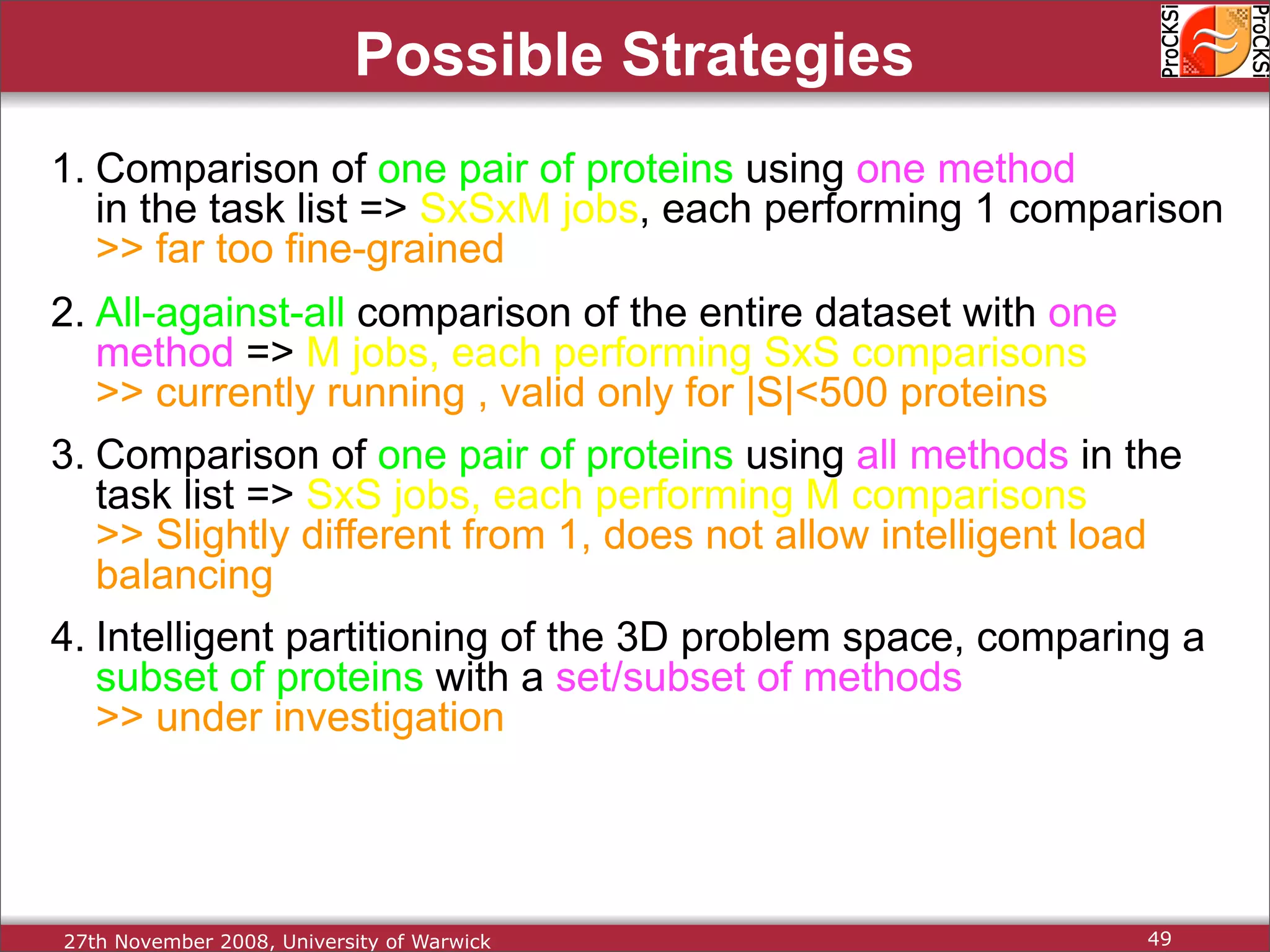
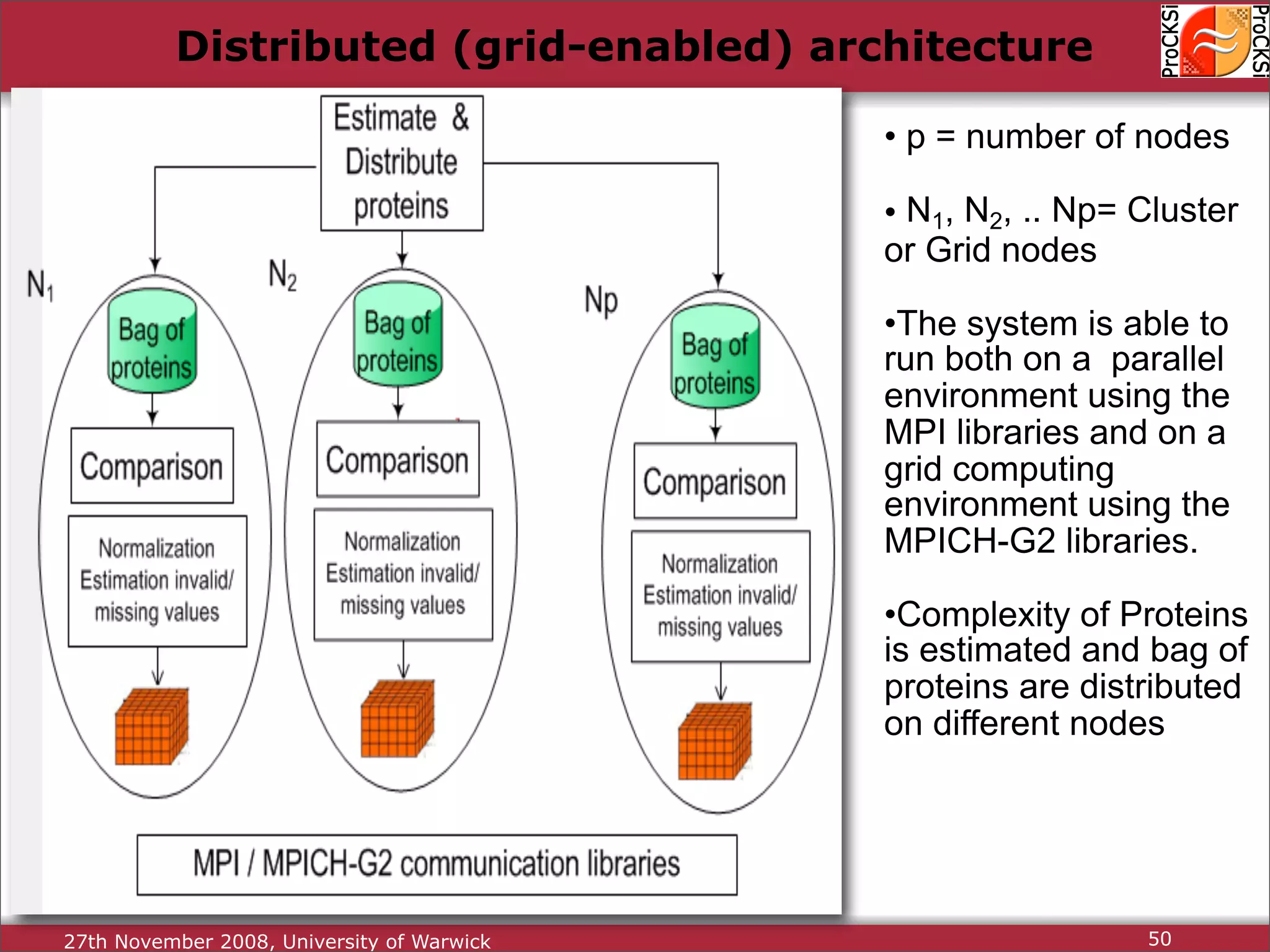
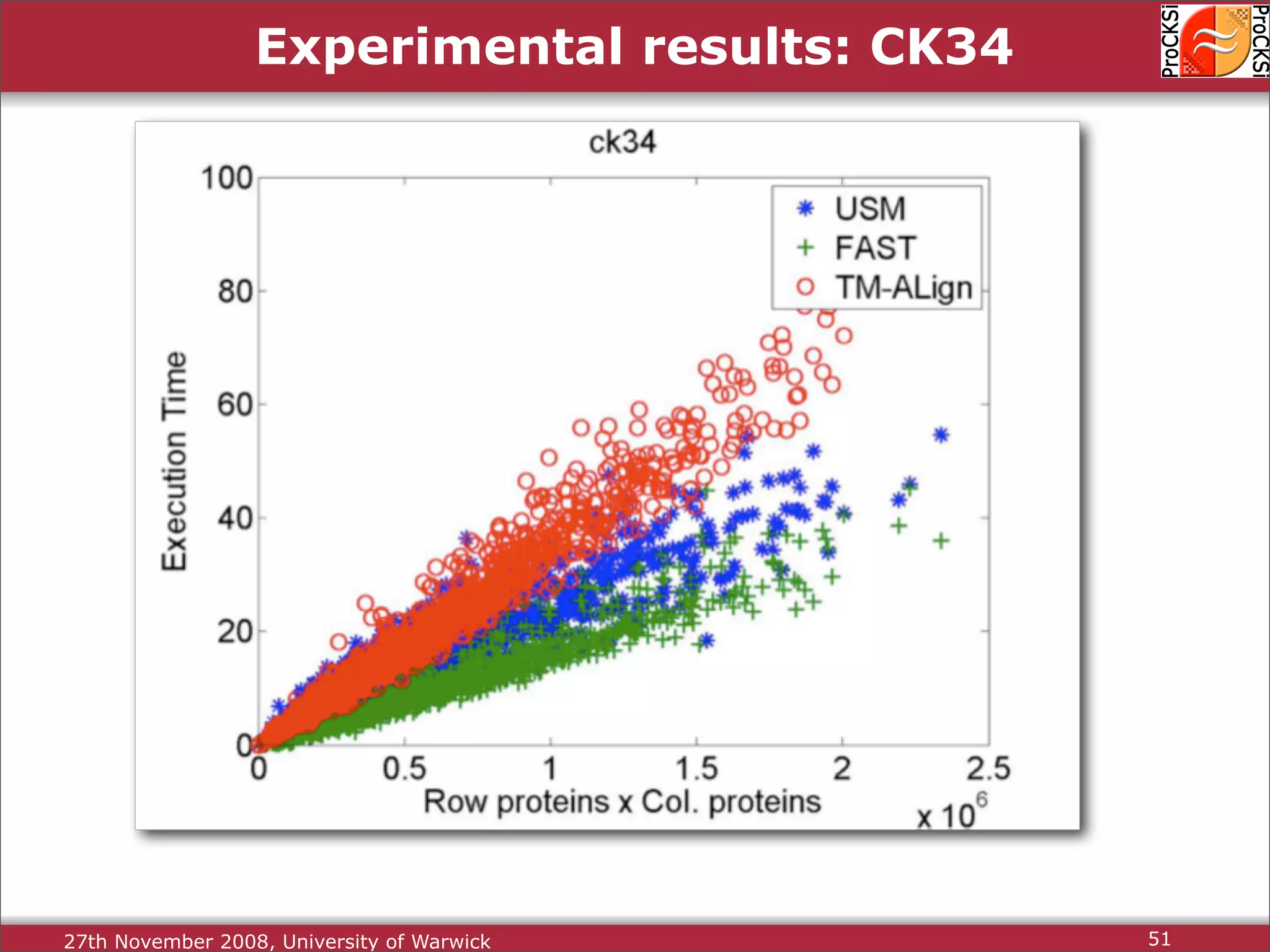
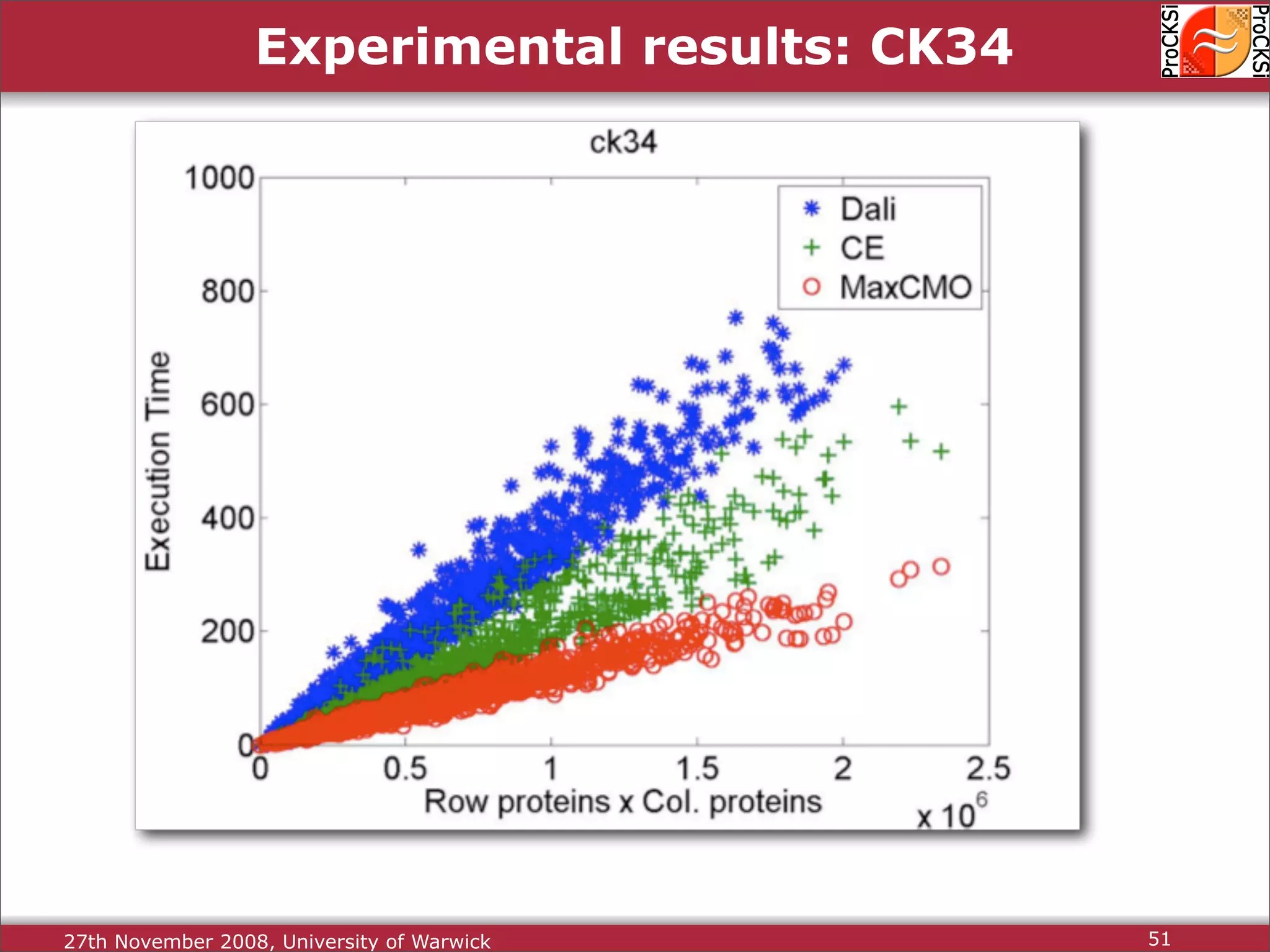
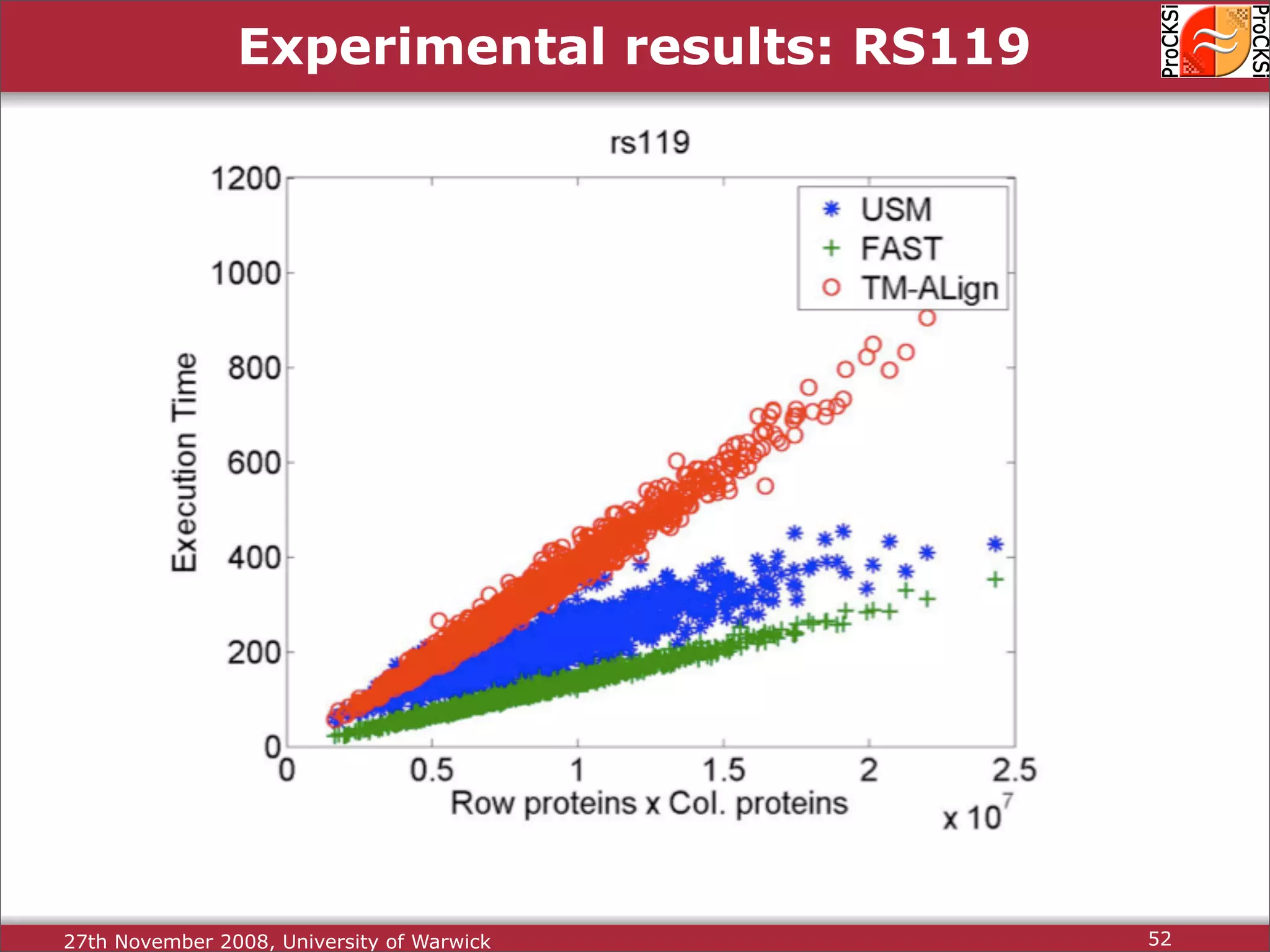
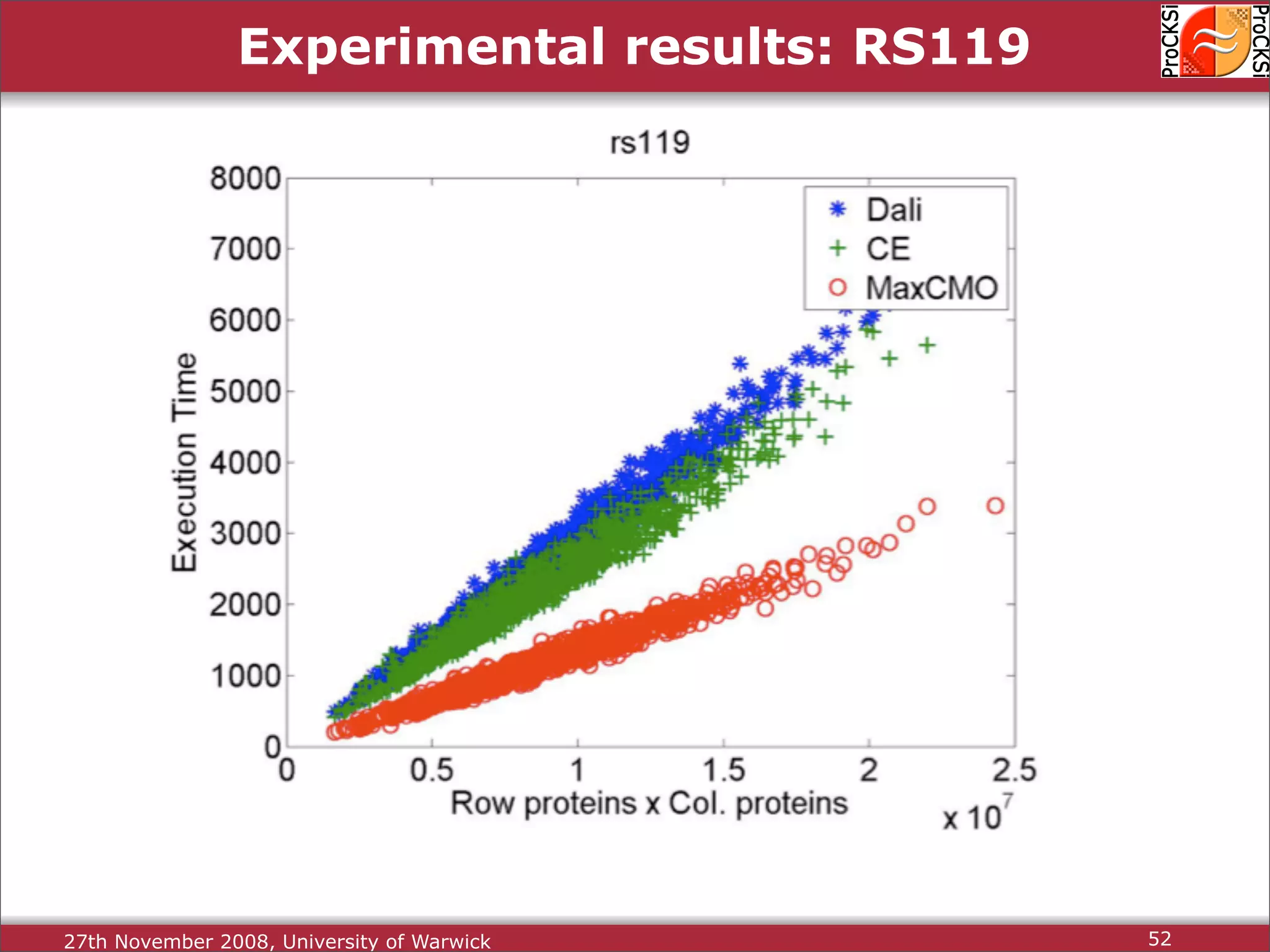
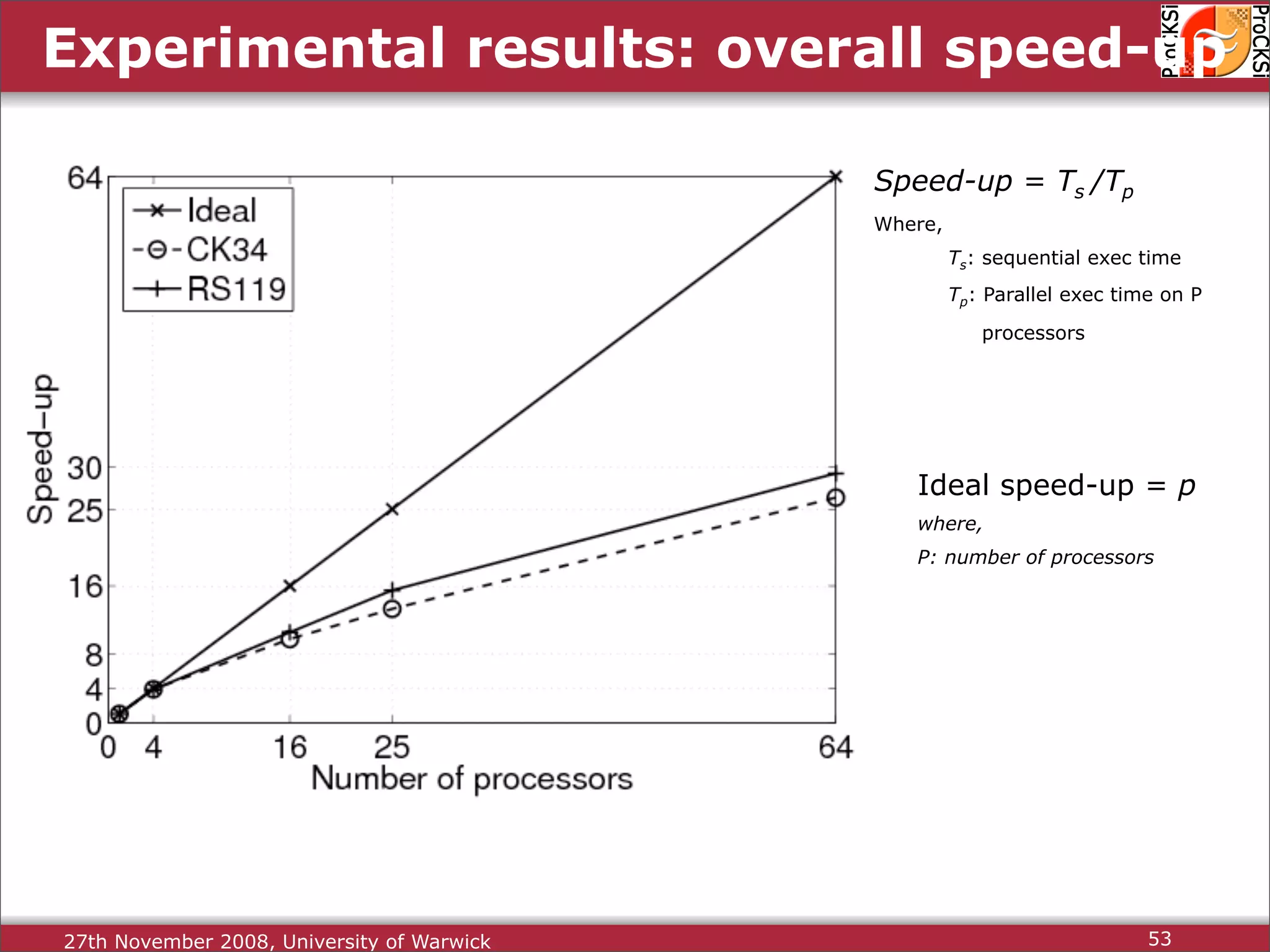
Overview of methodologies in distributed computing for protein comparison, emphasizing performance and efficiency.
Summary of ProCKSI's functionalities and ongoing expansions, inviting contributions for further improvements.
Acknowledgments for contributions and a list of relevant literature supporting the study.