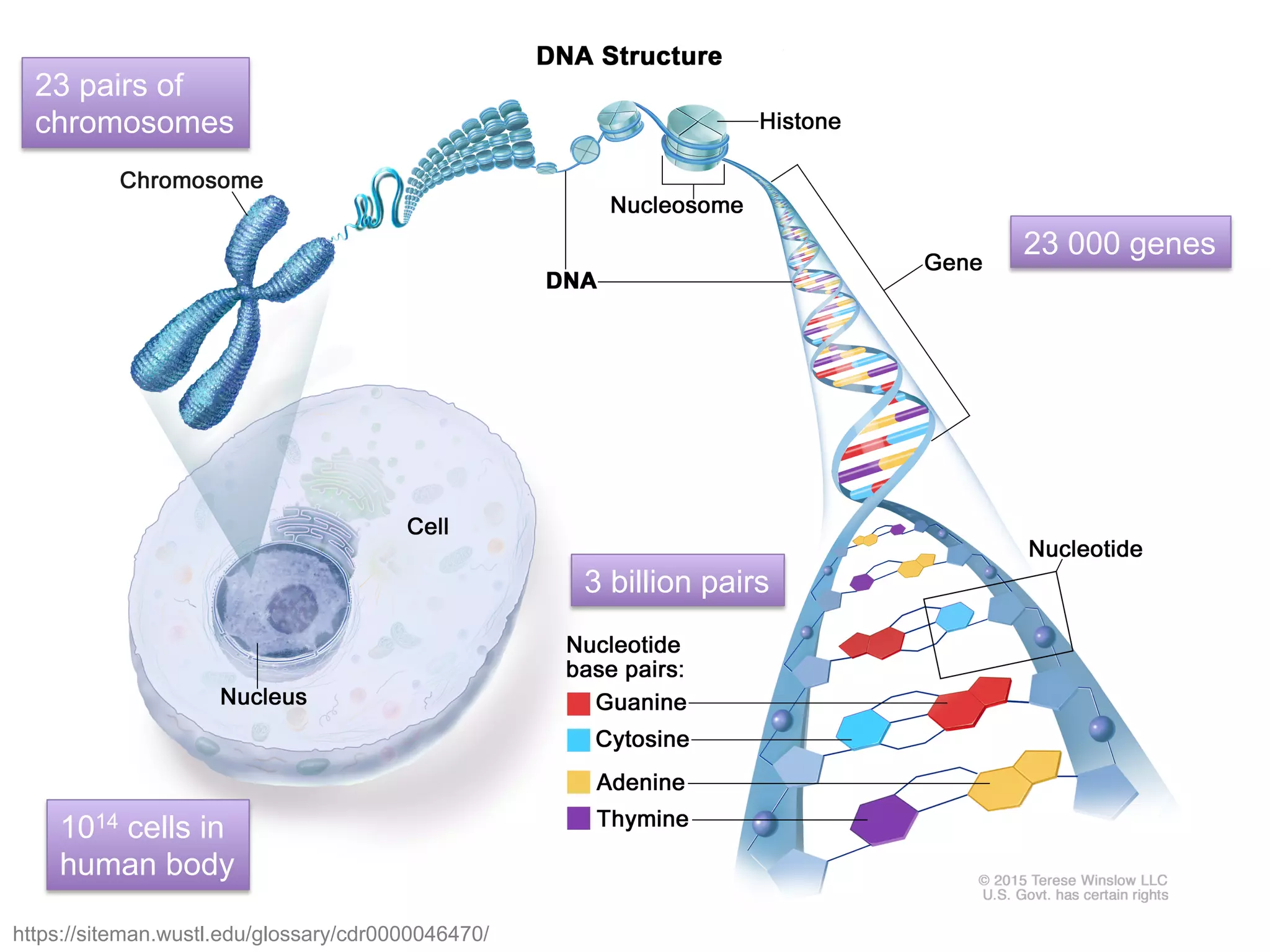
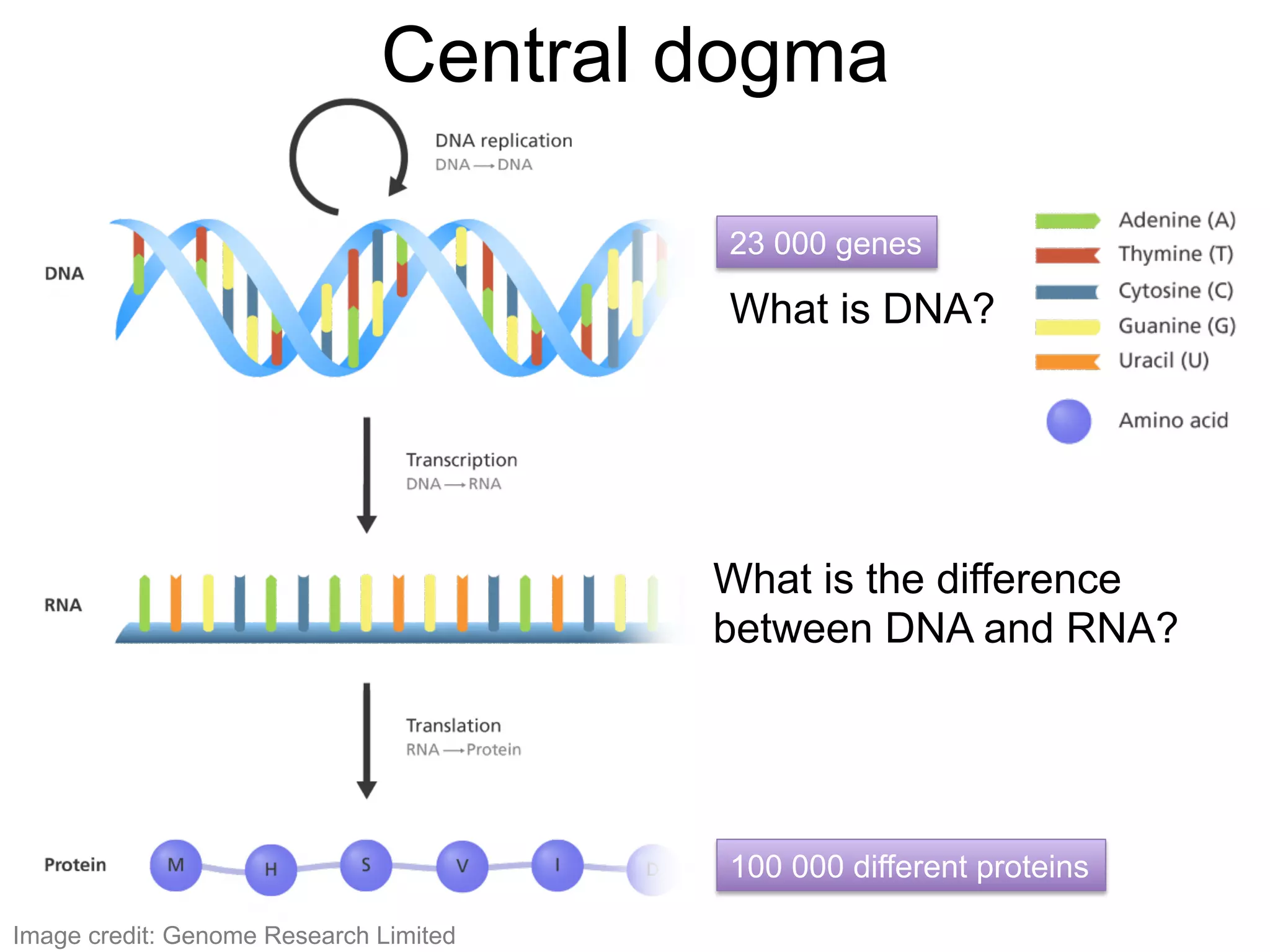
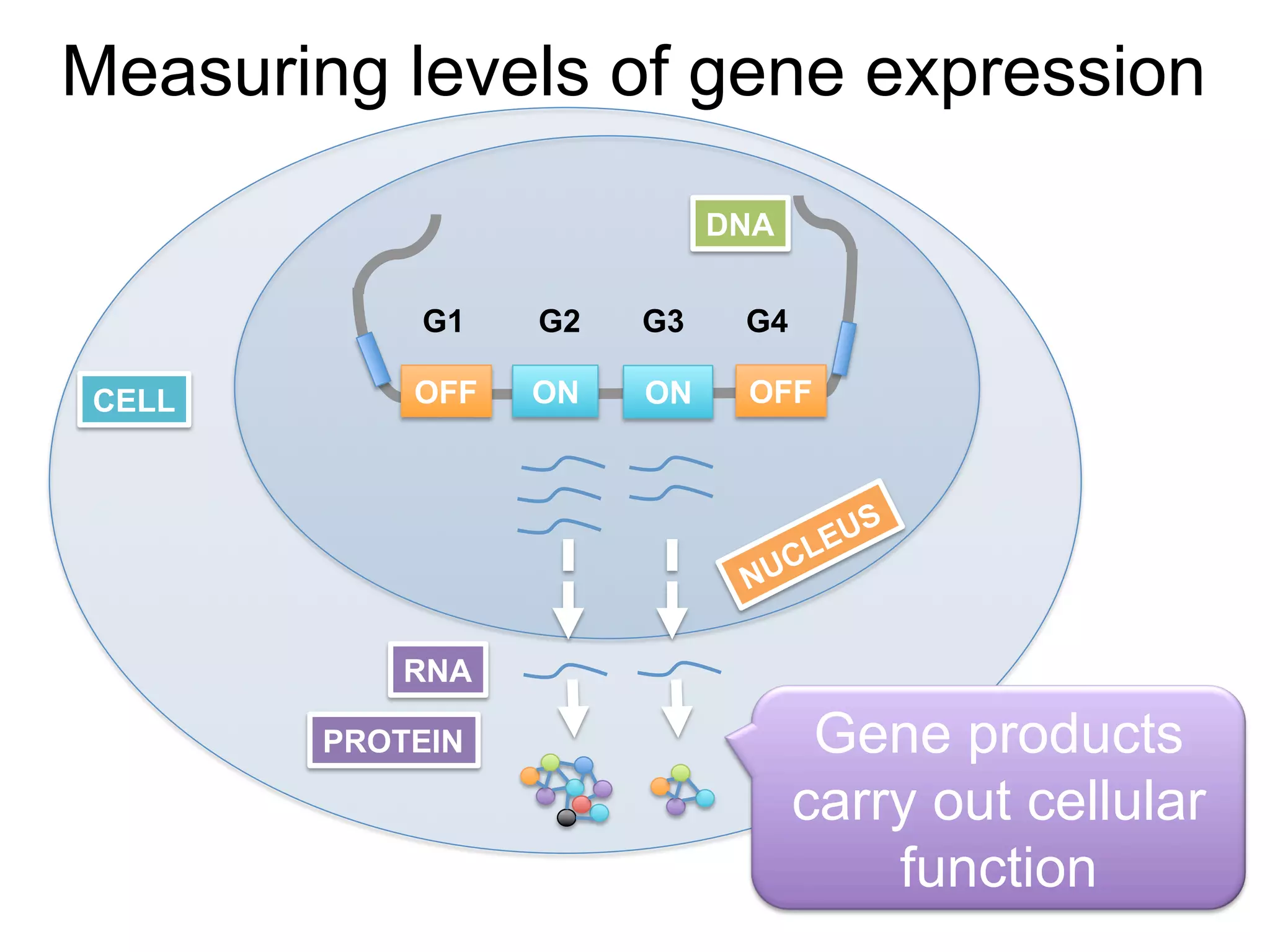
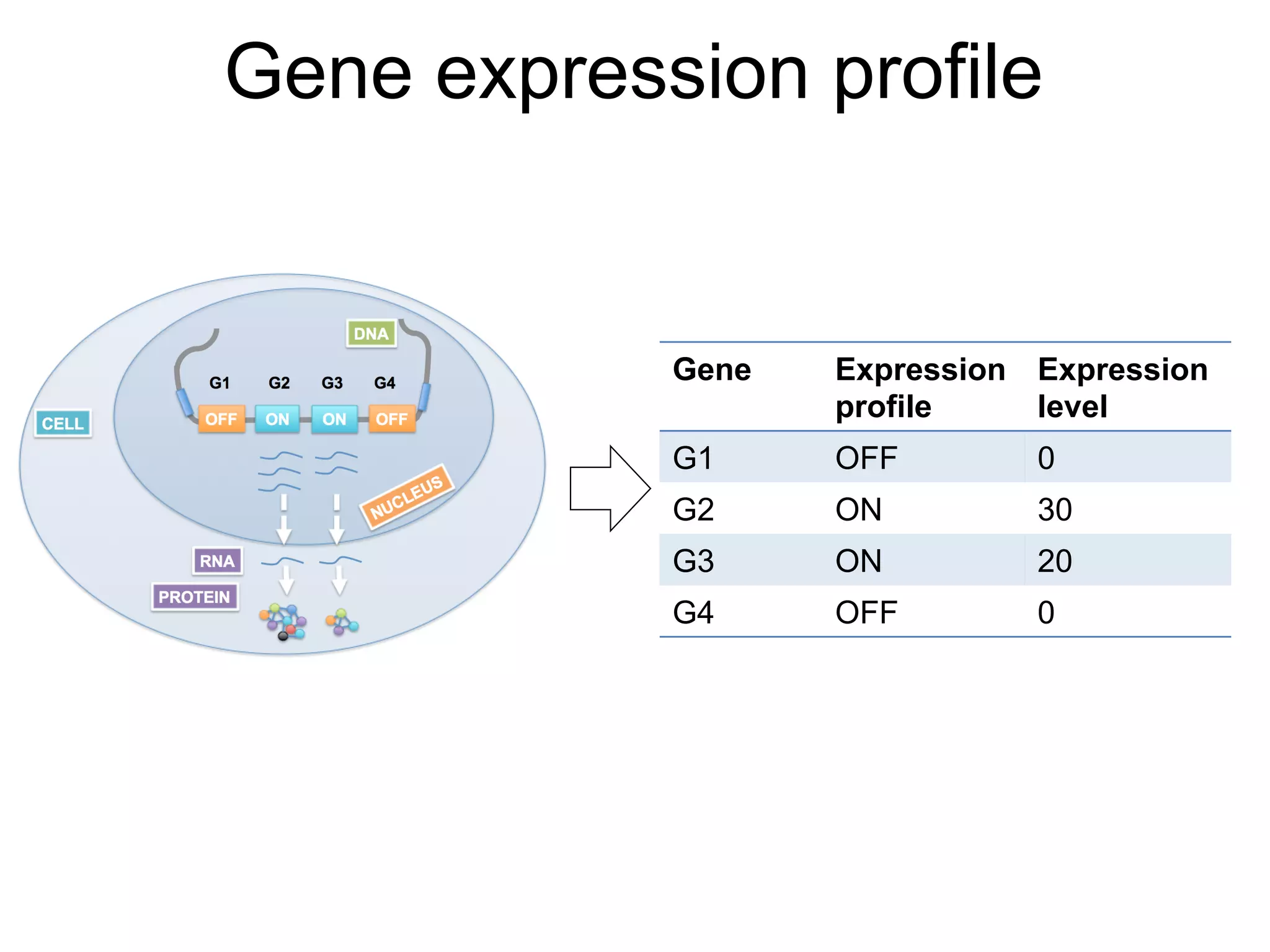
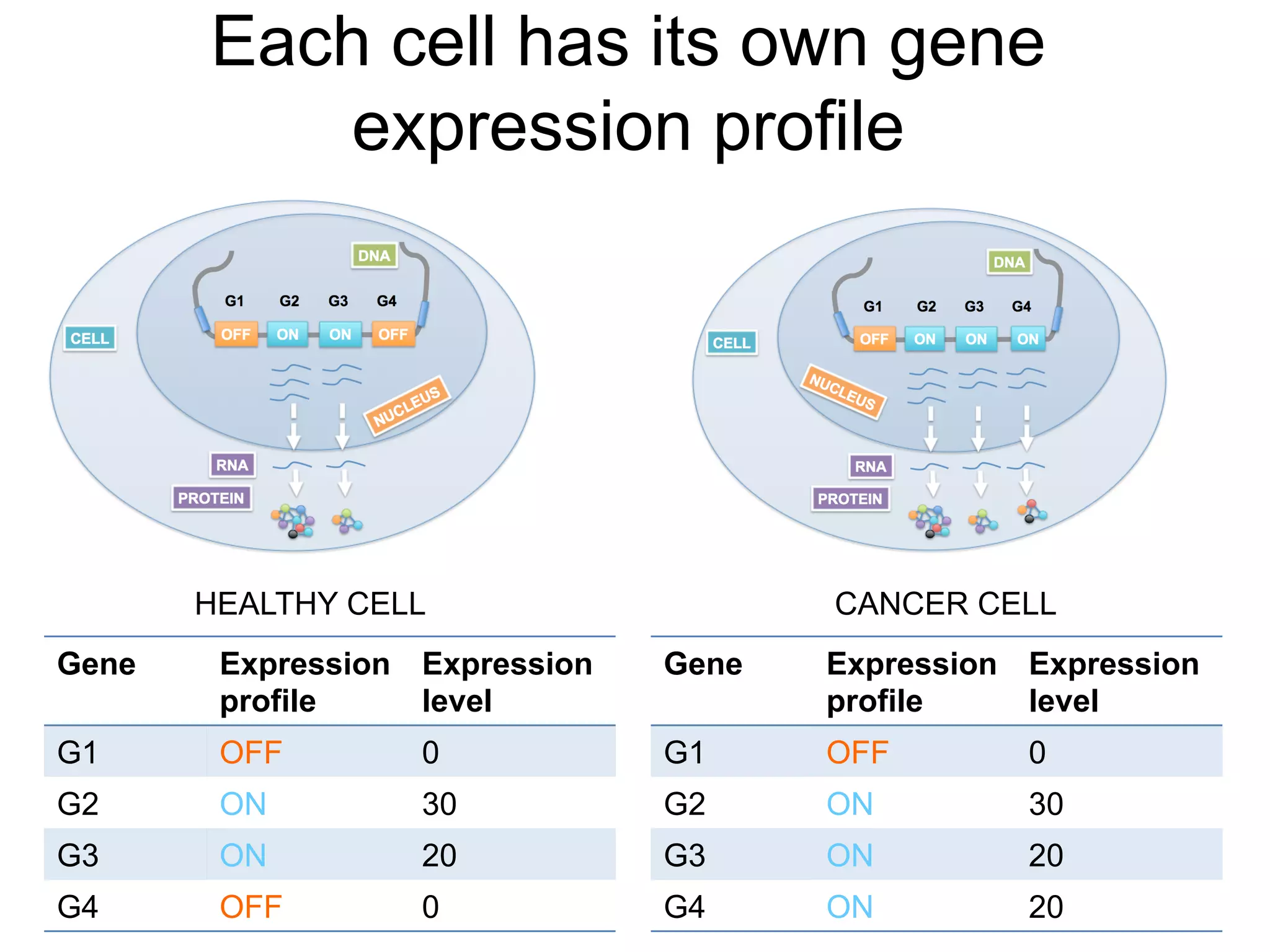
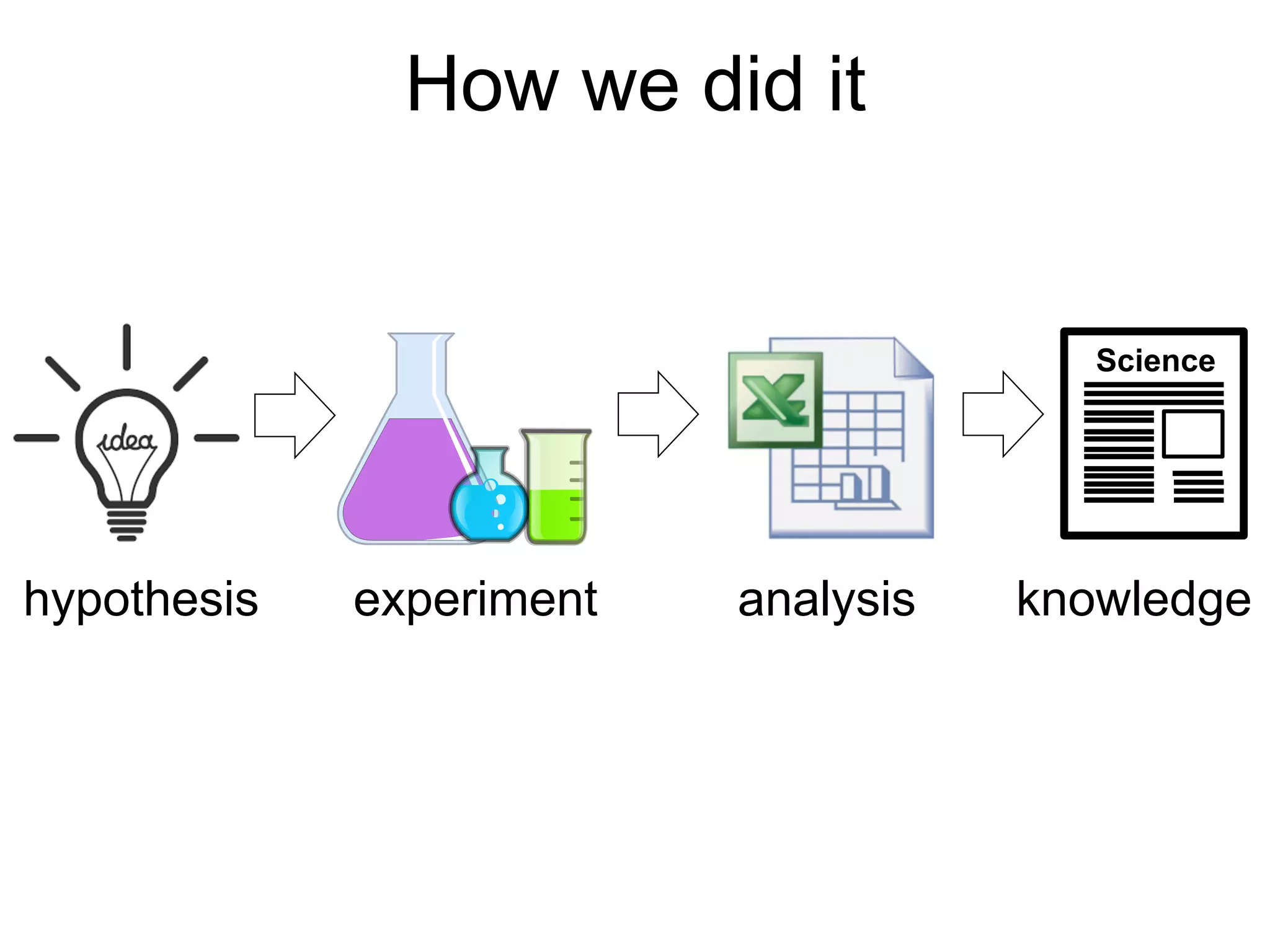
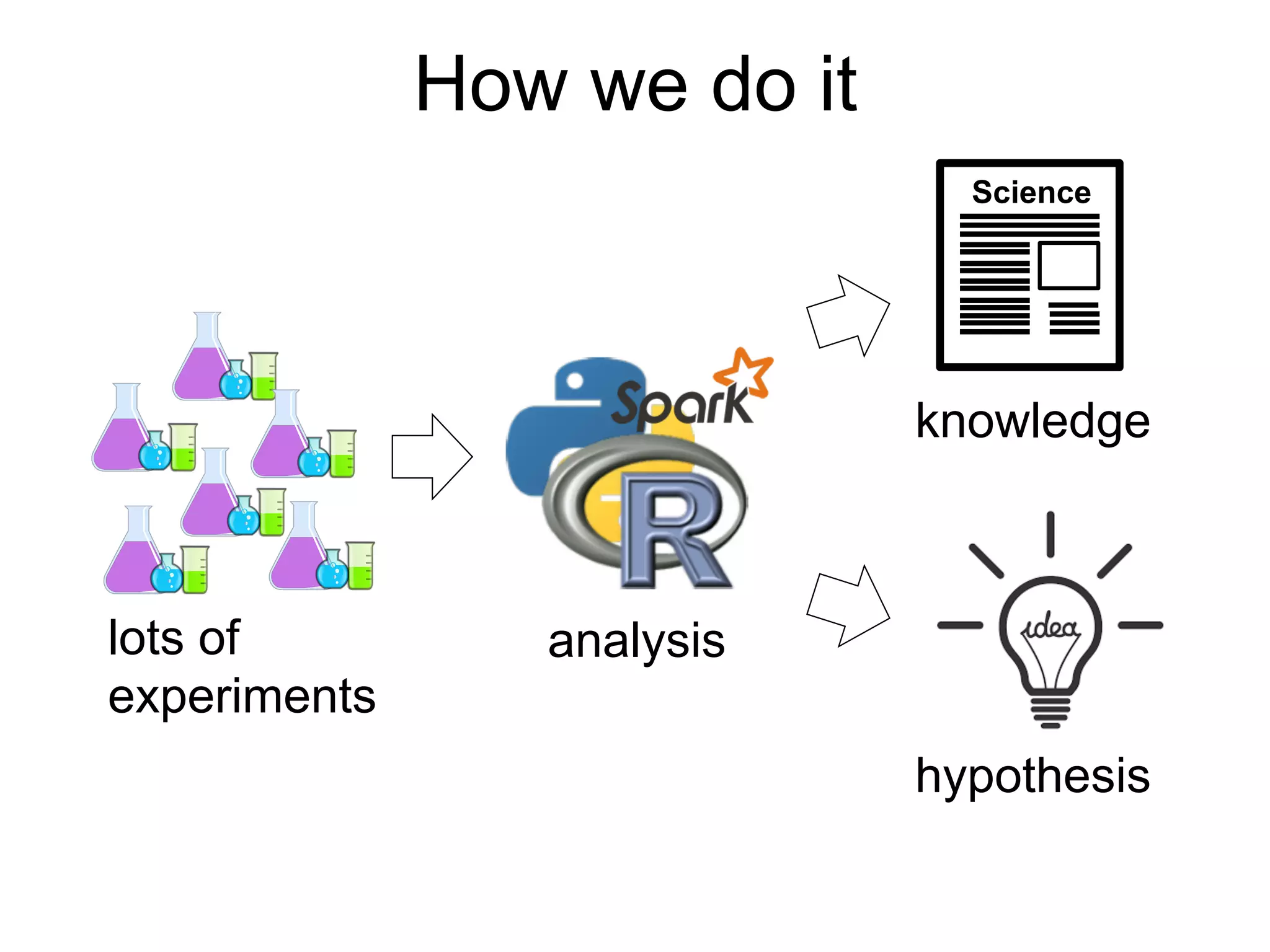
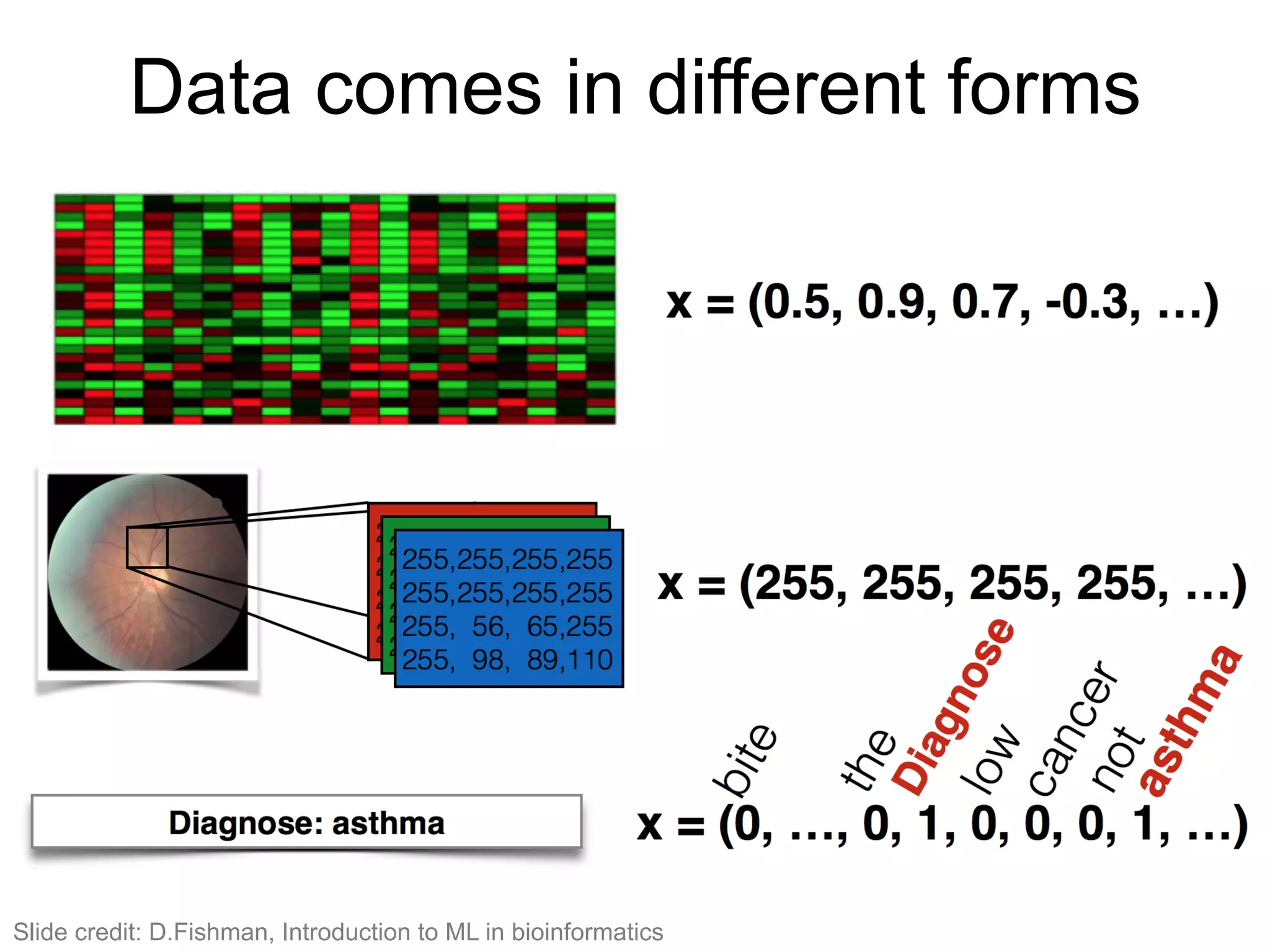
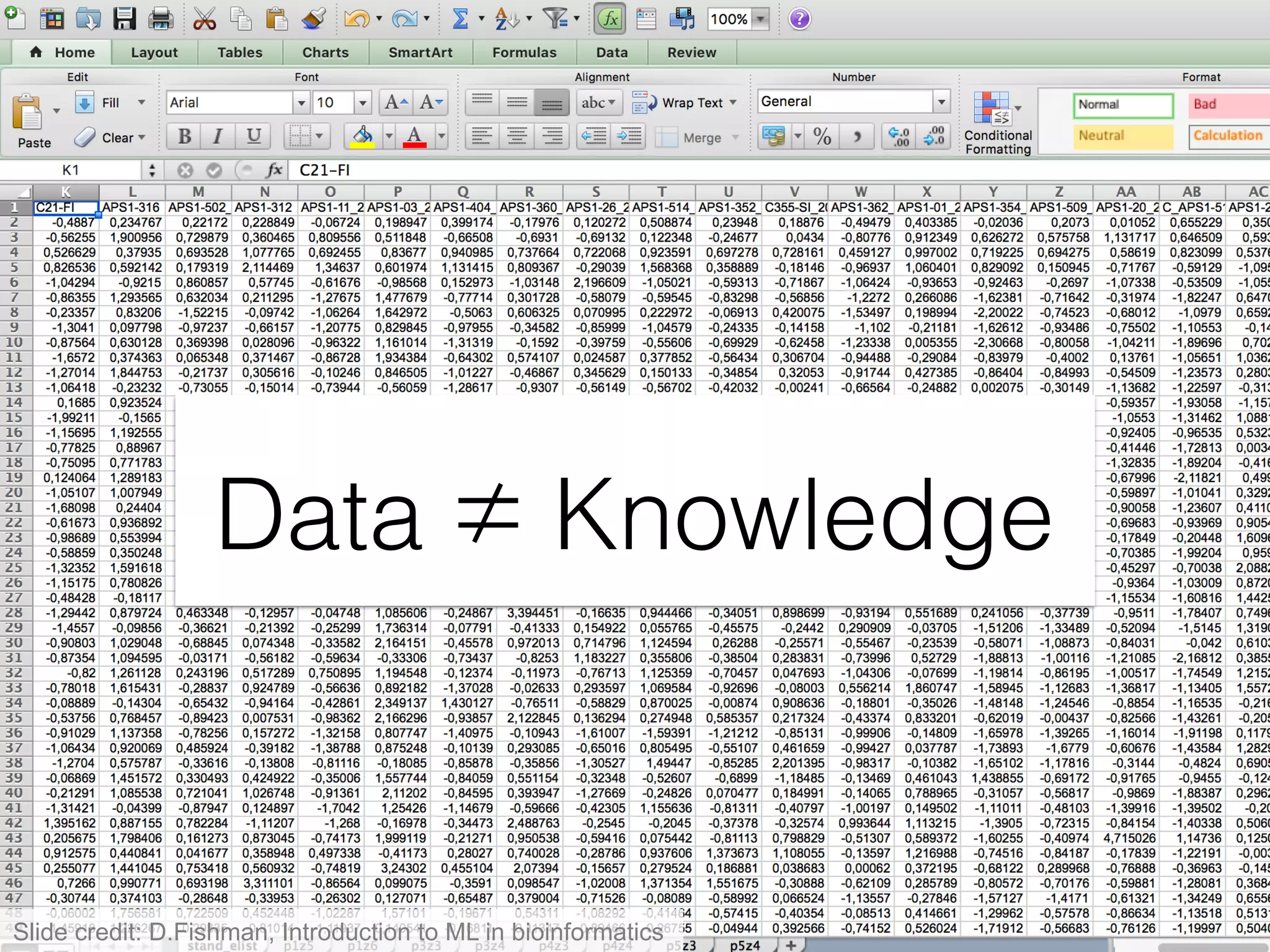
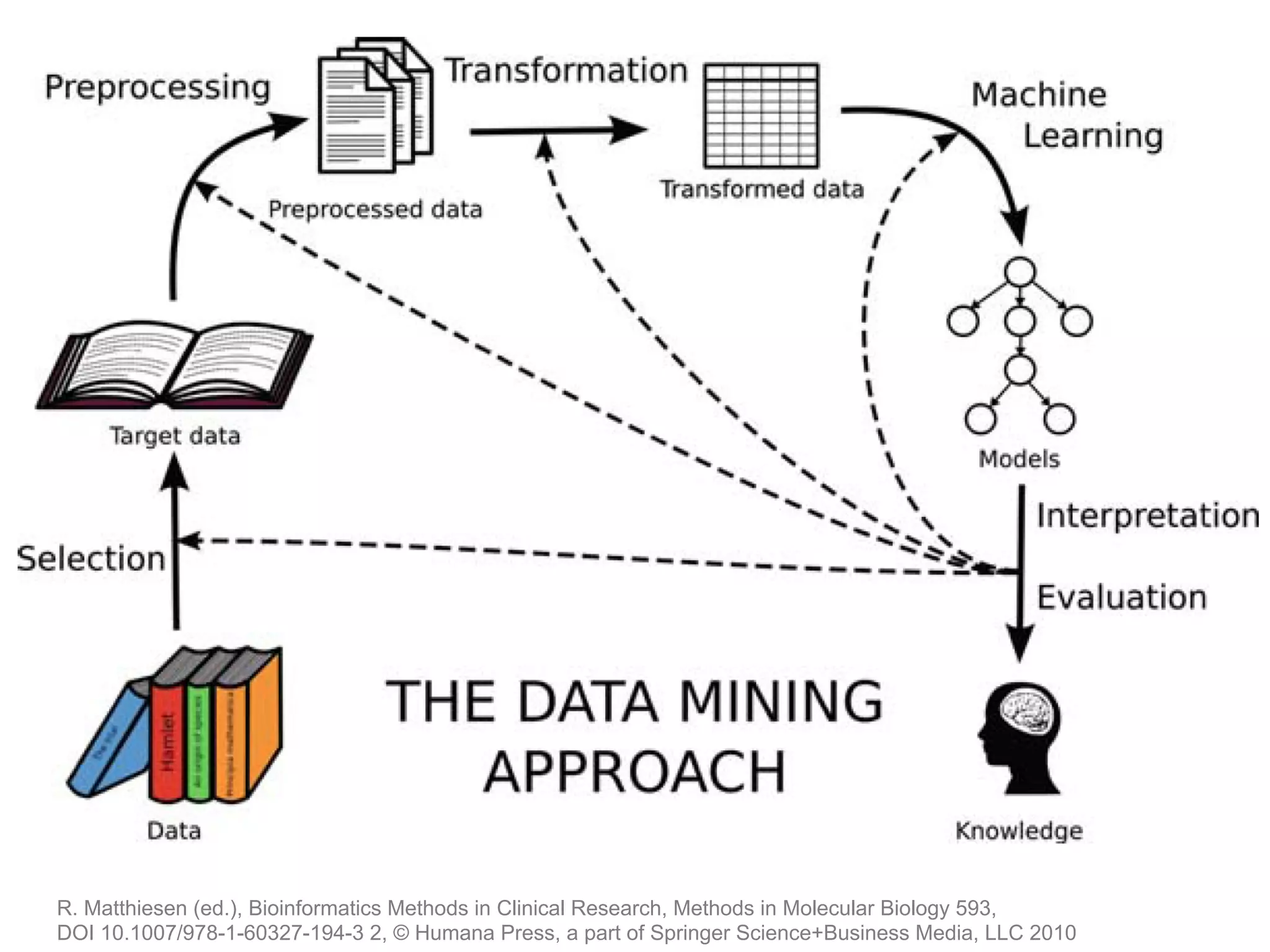
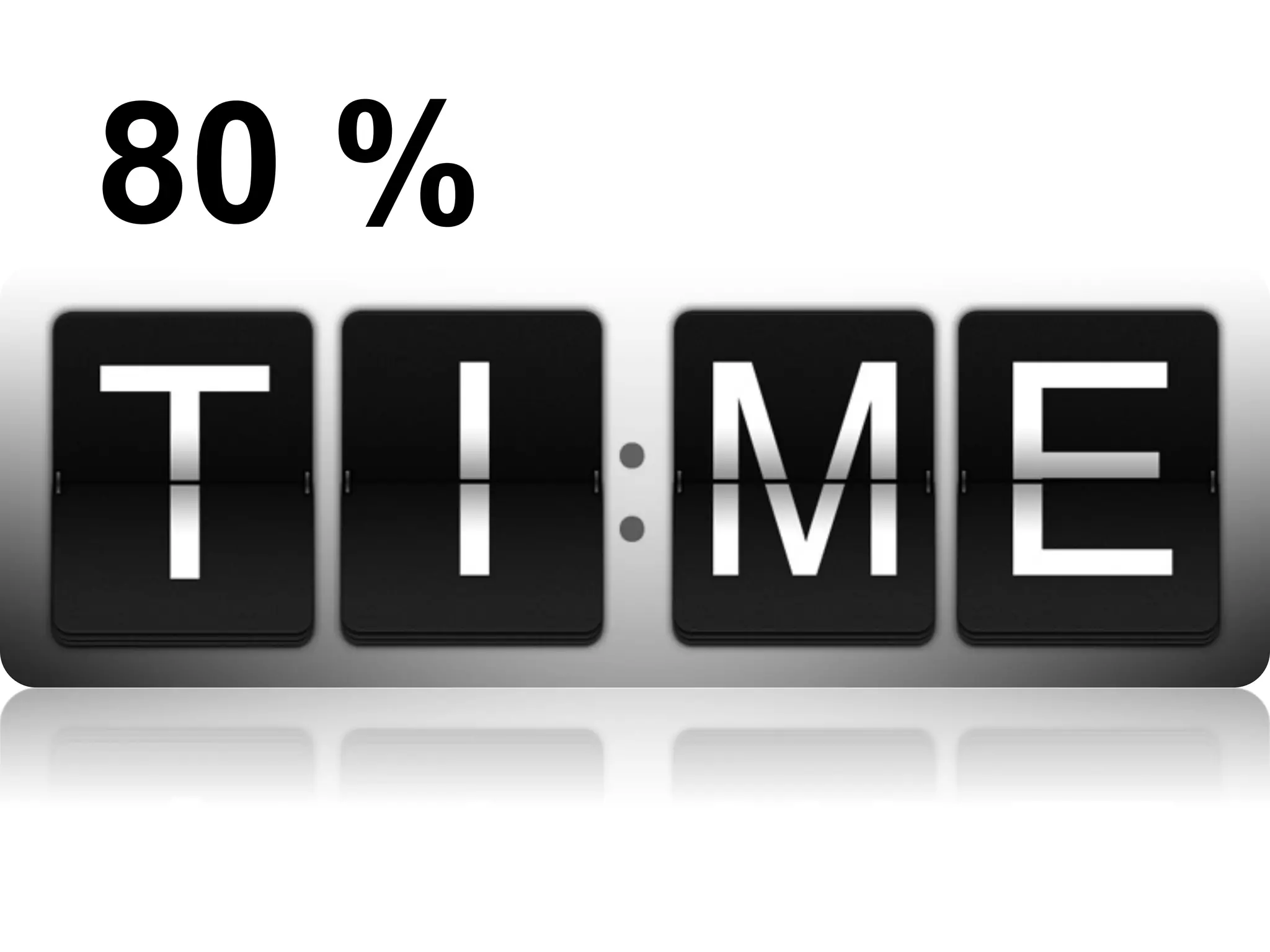
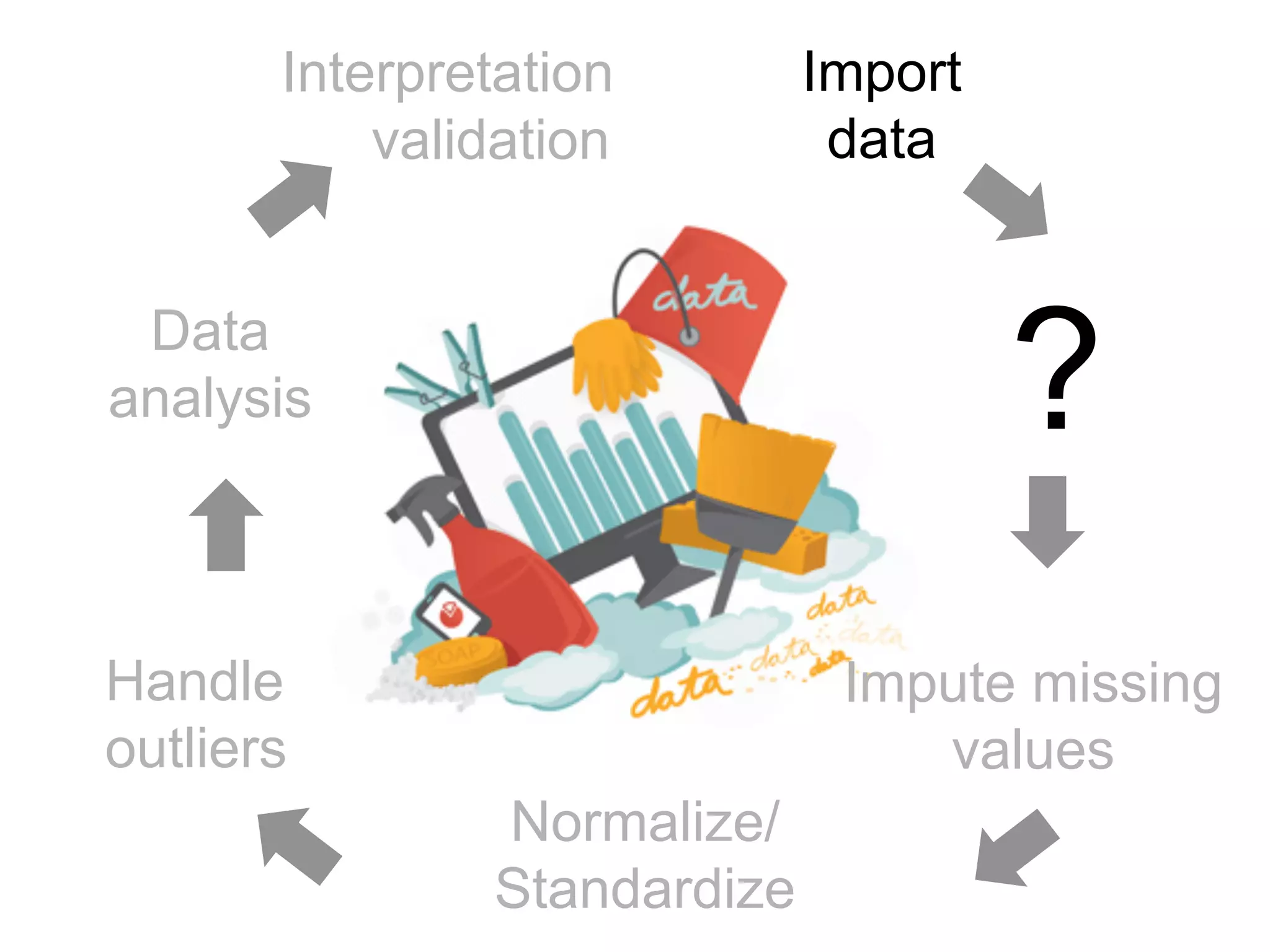
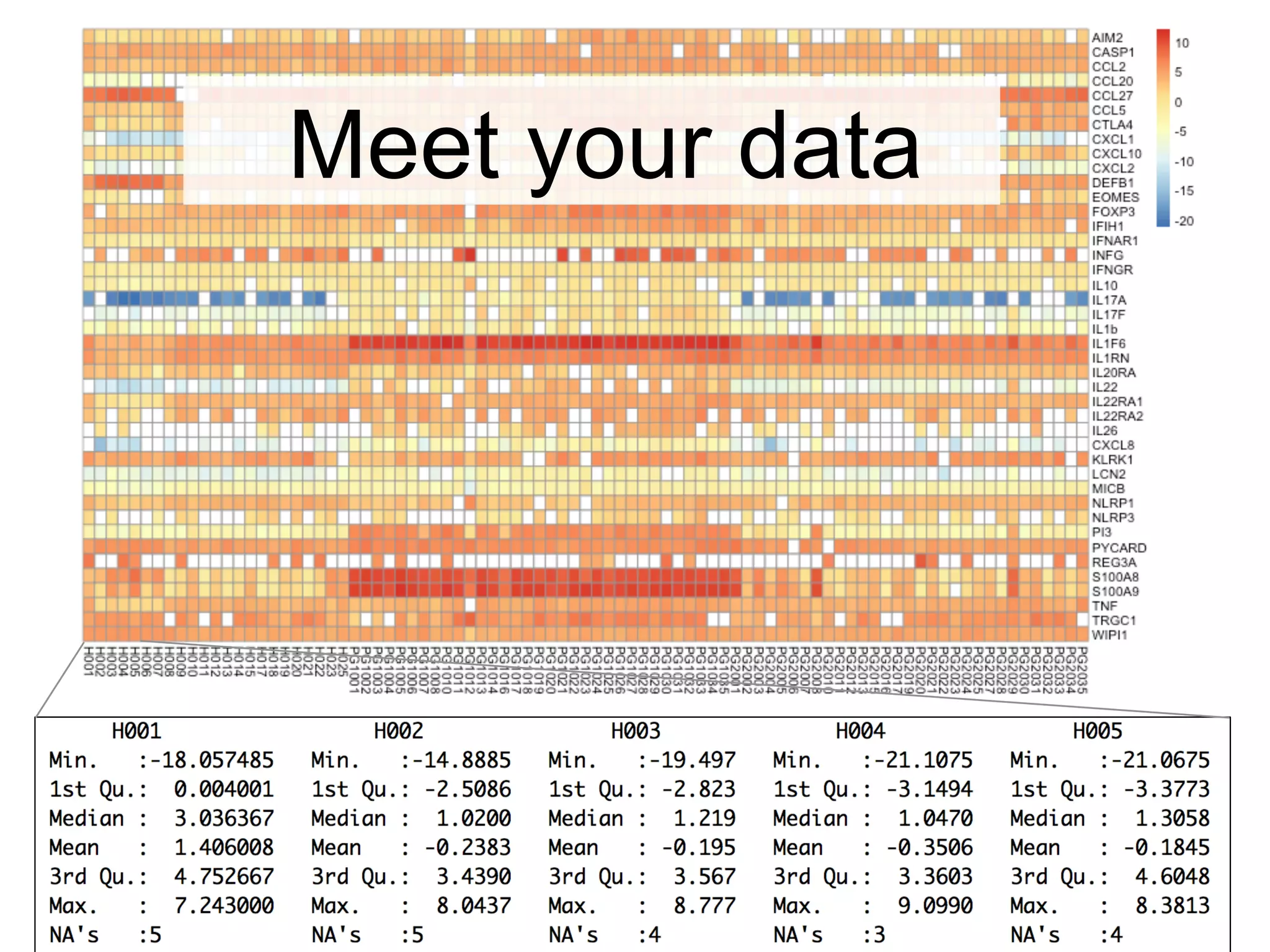
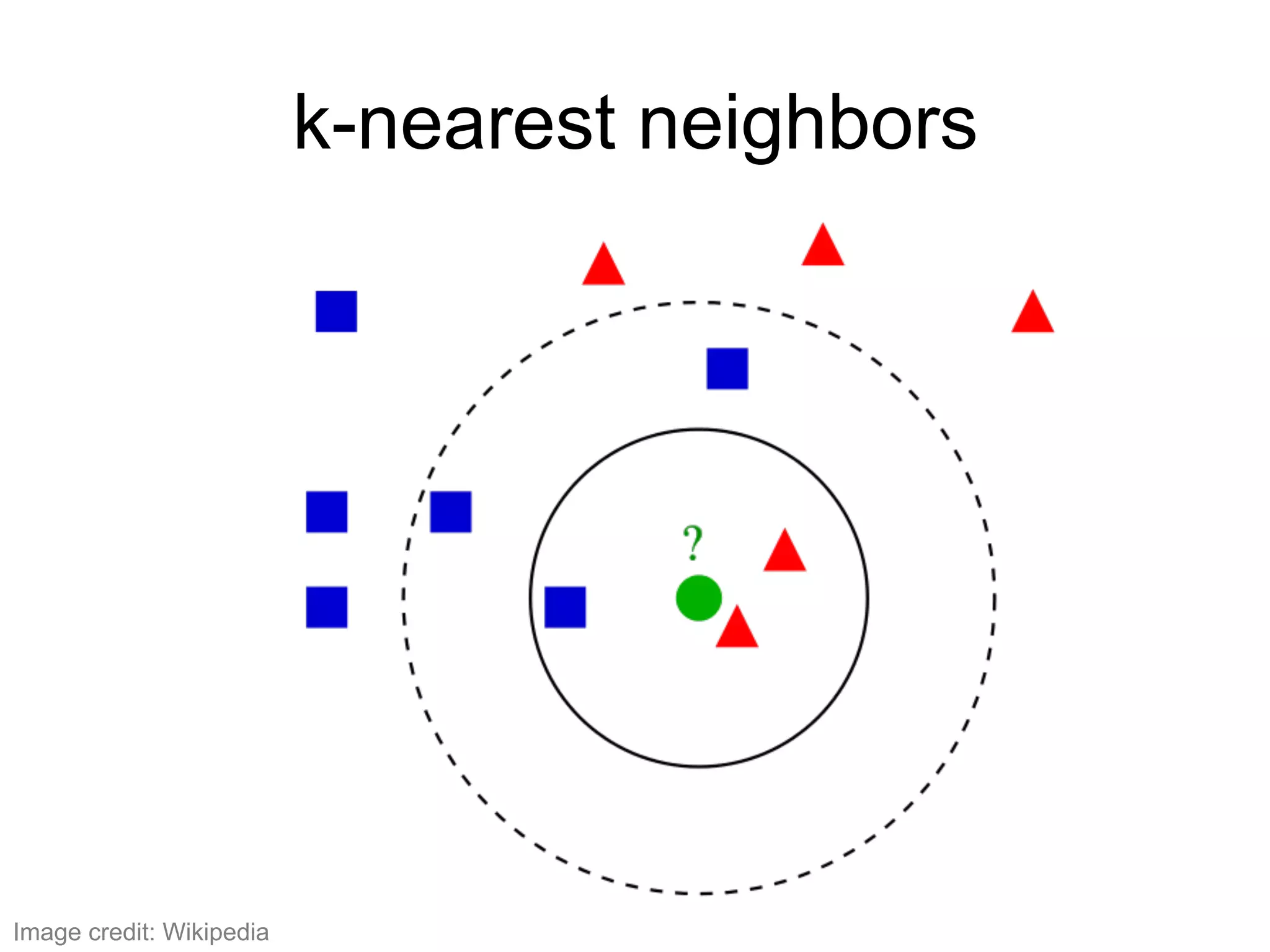
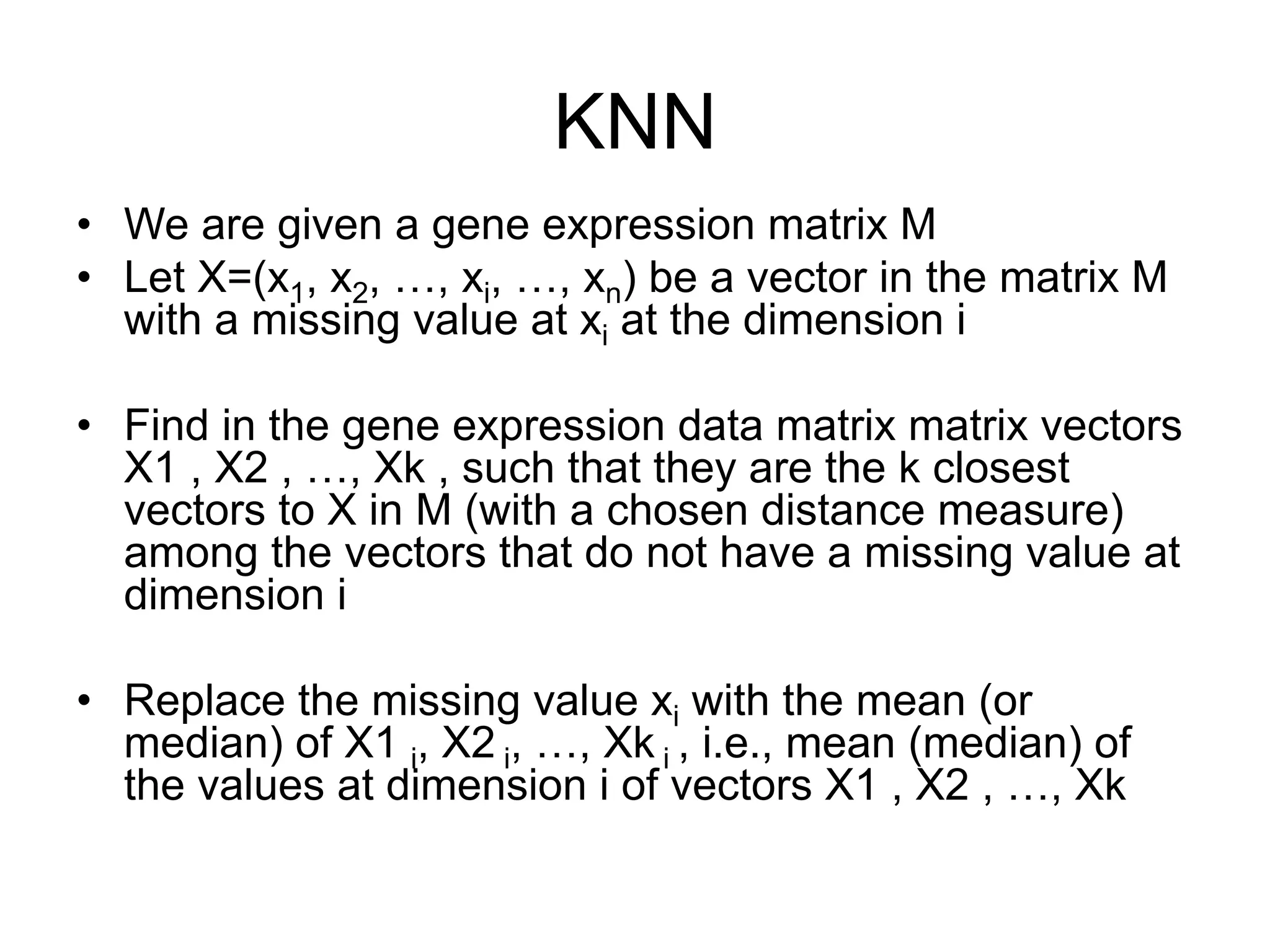
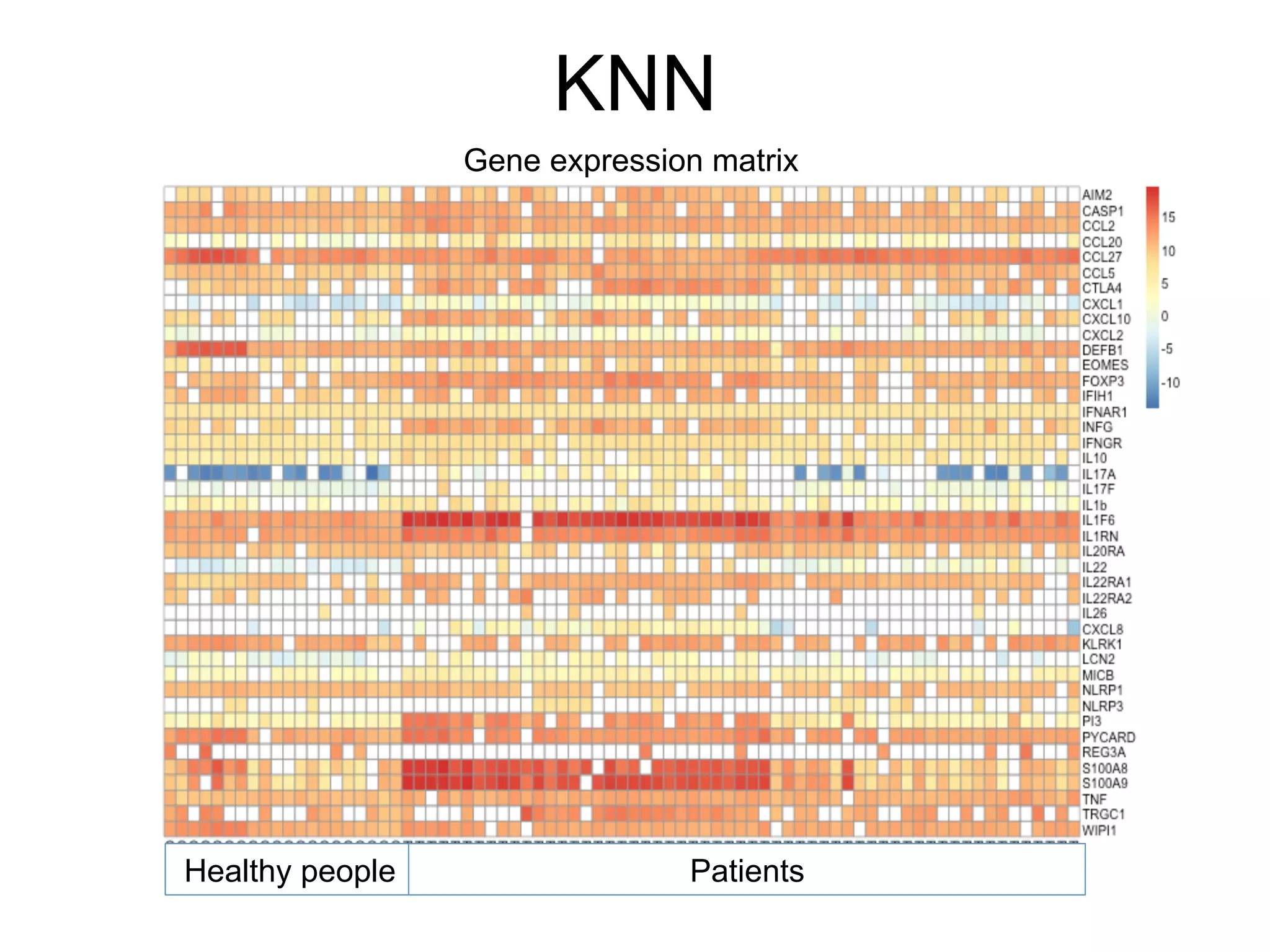
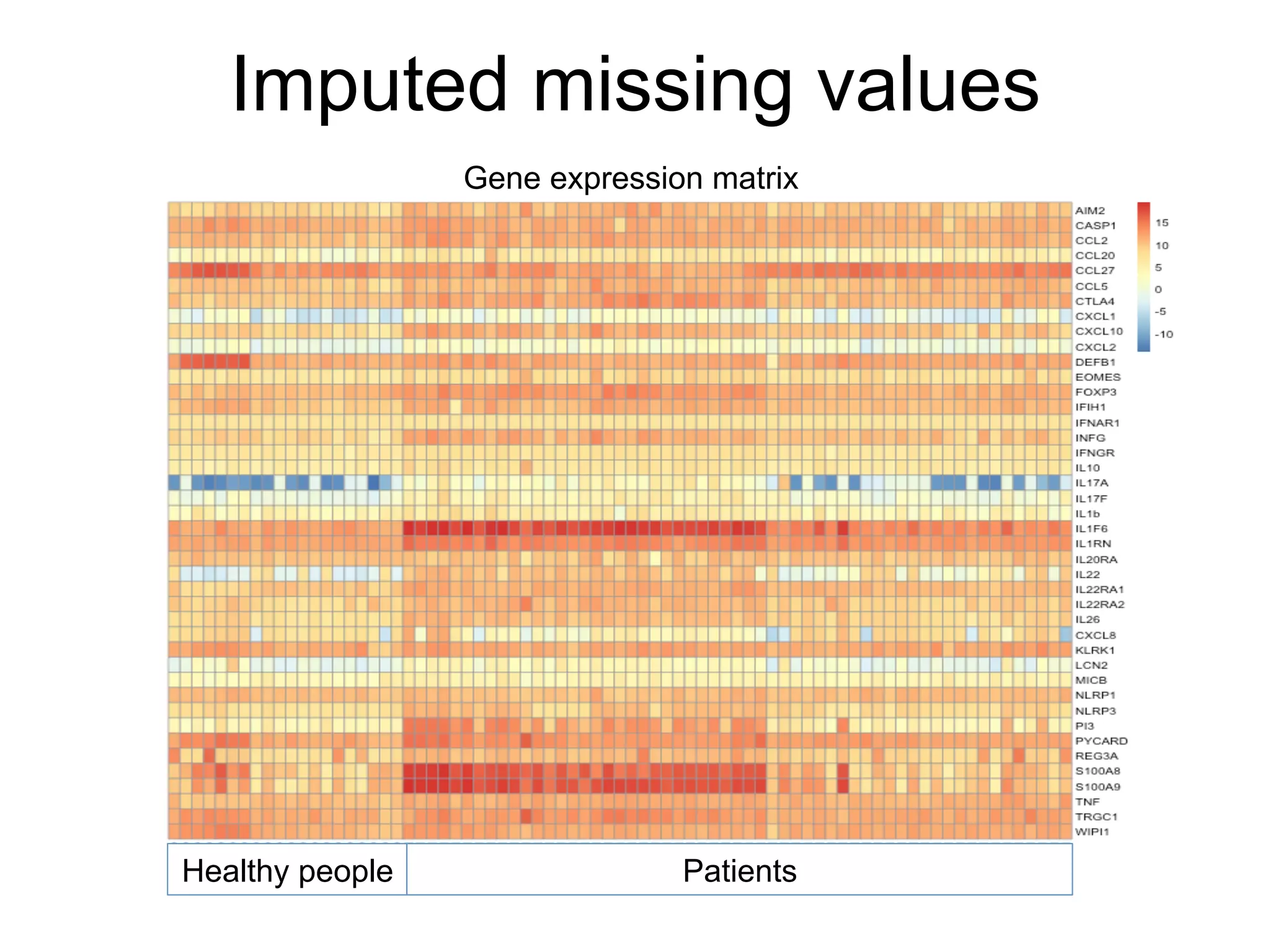
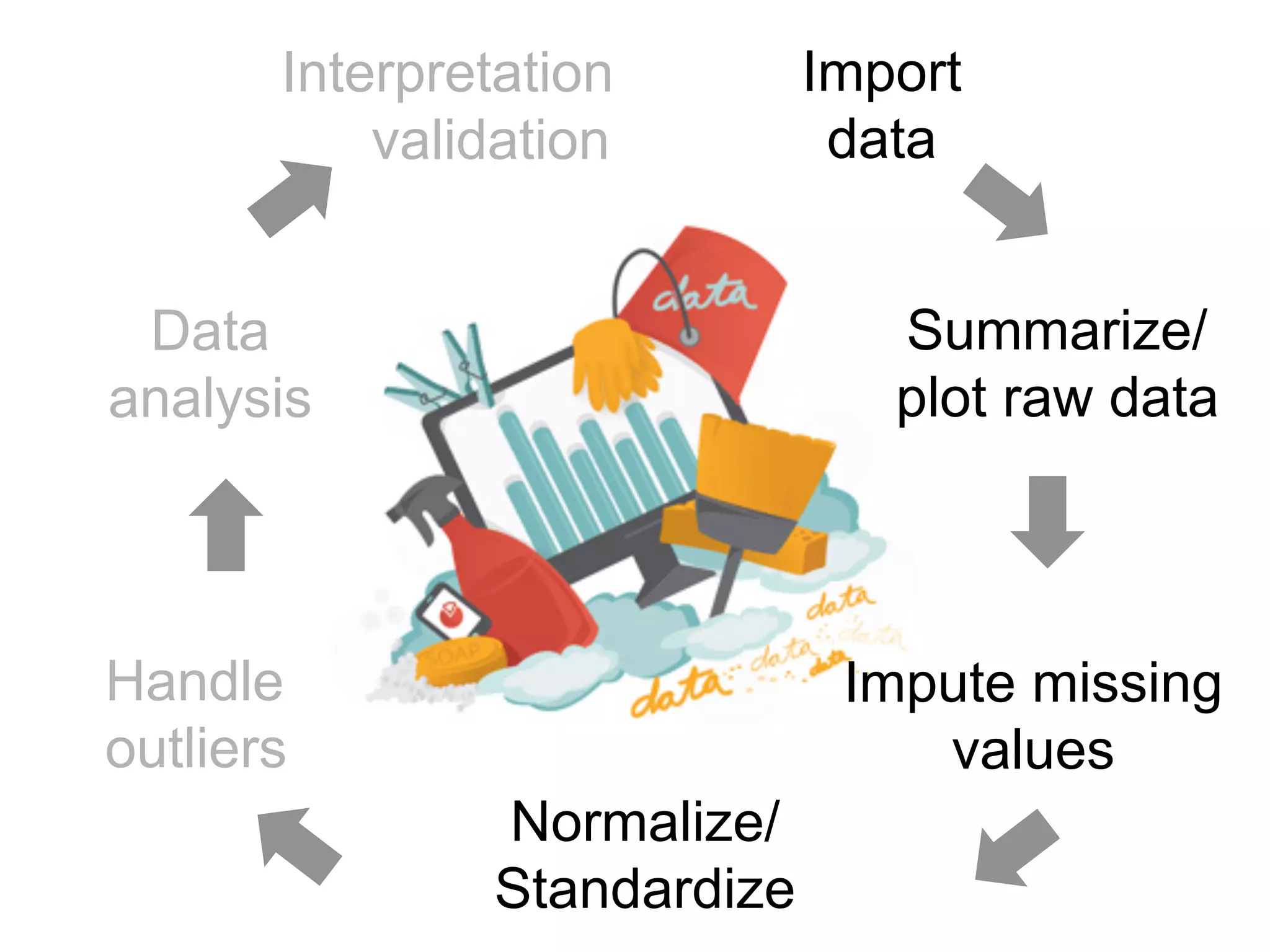
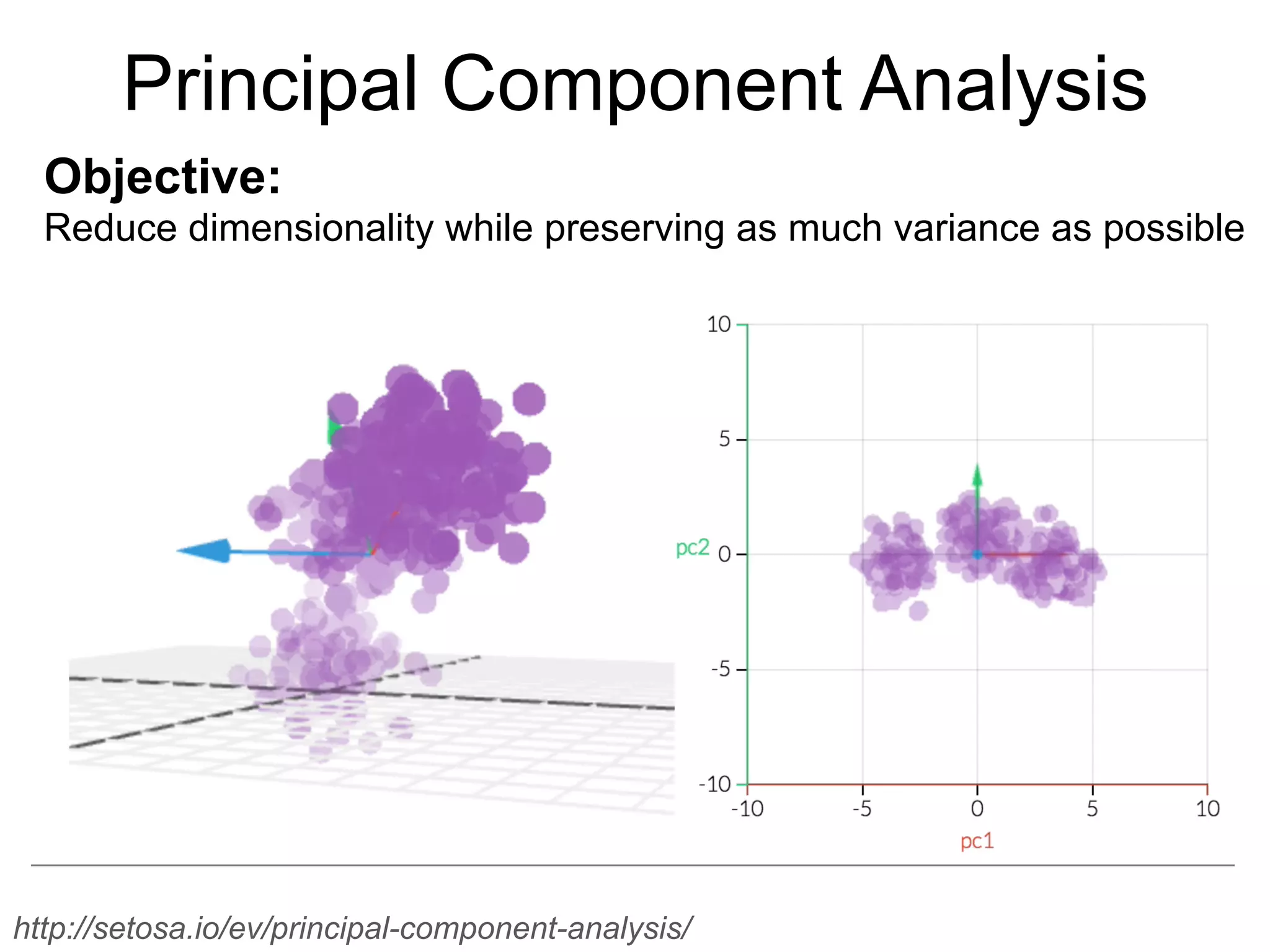
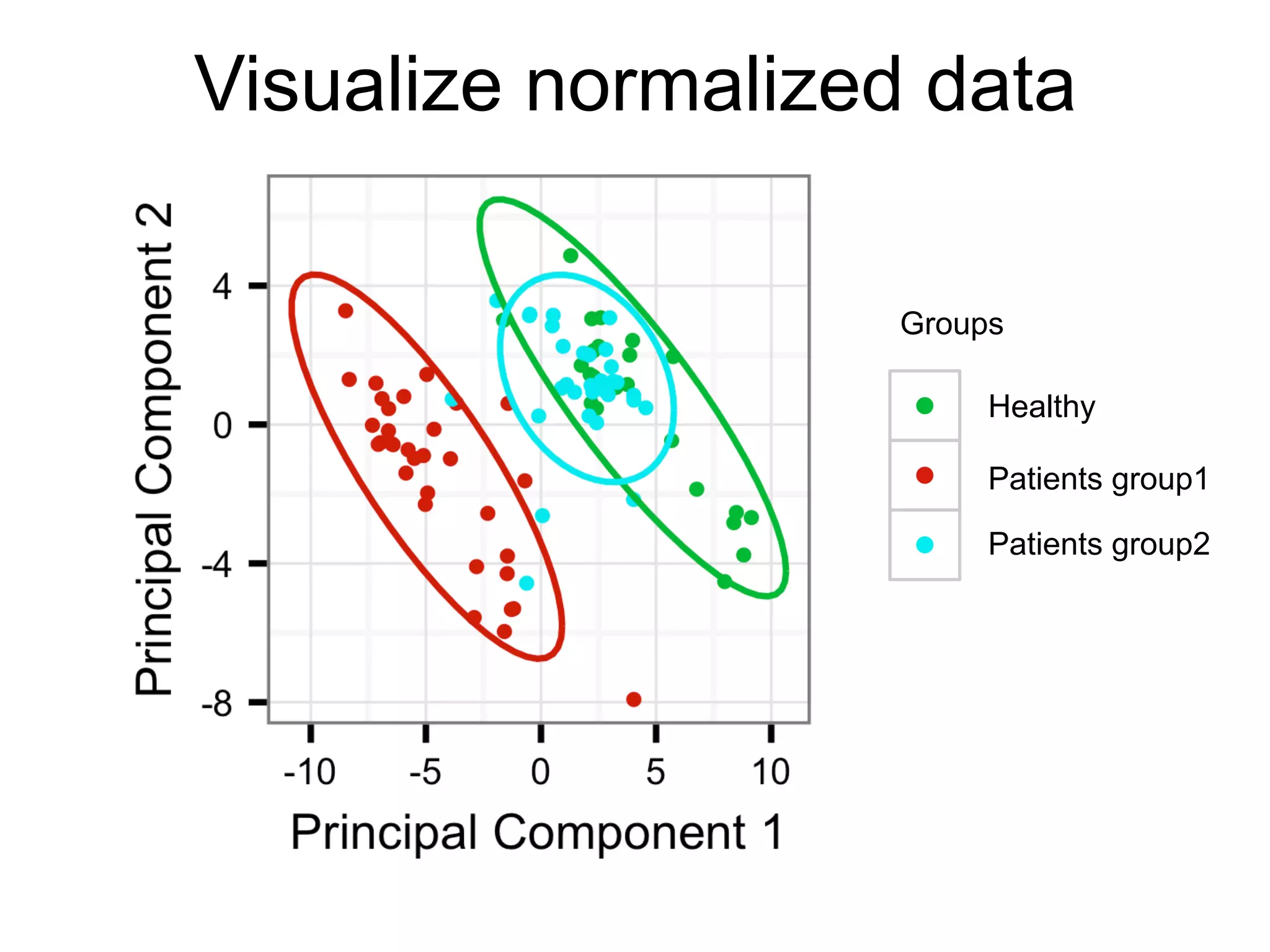
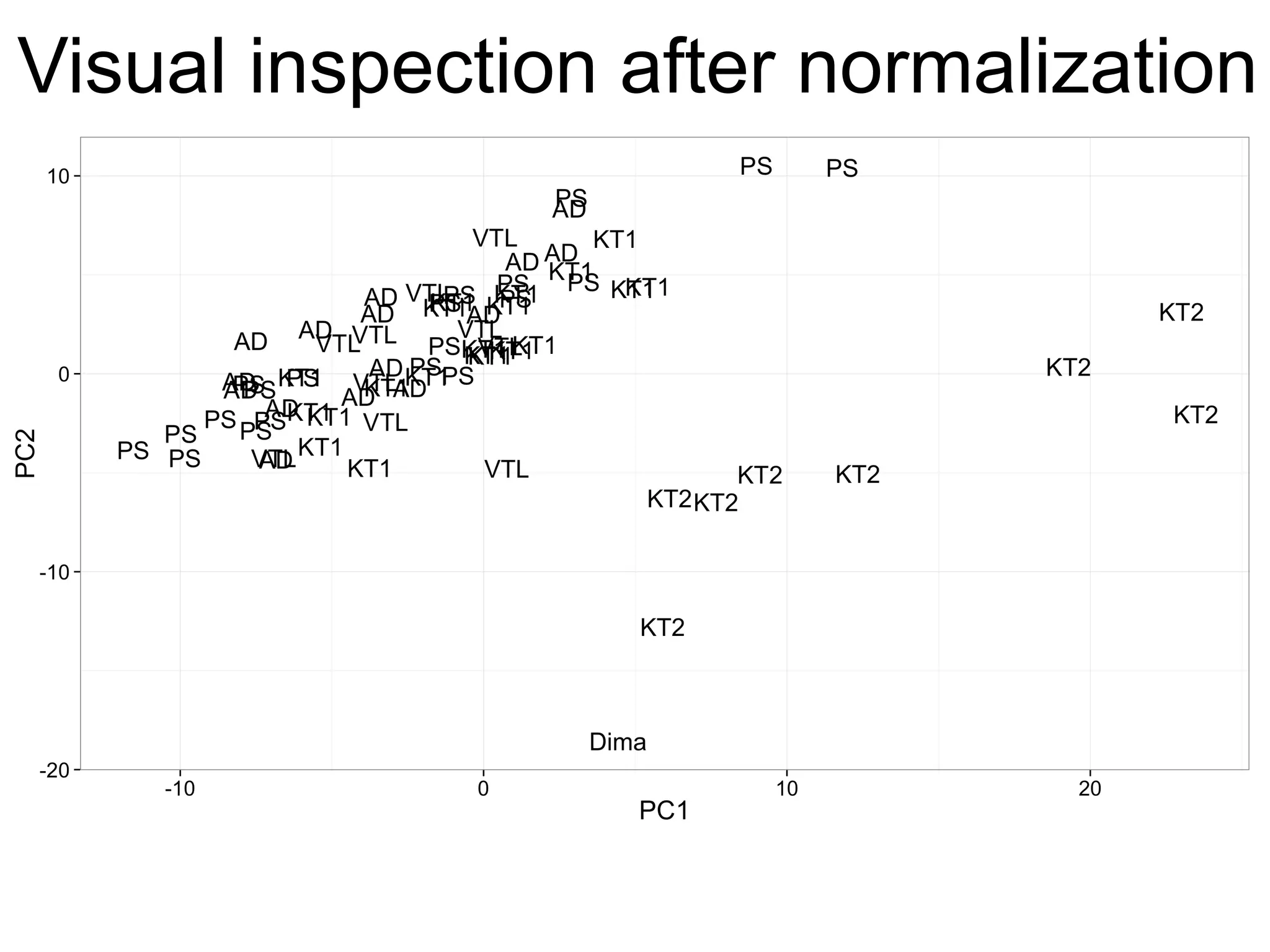
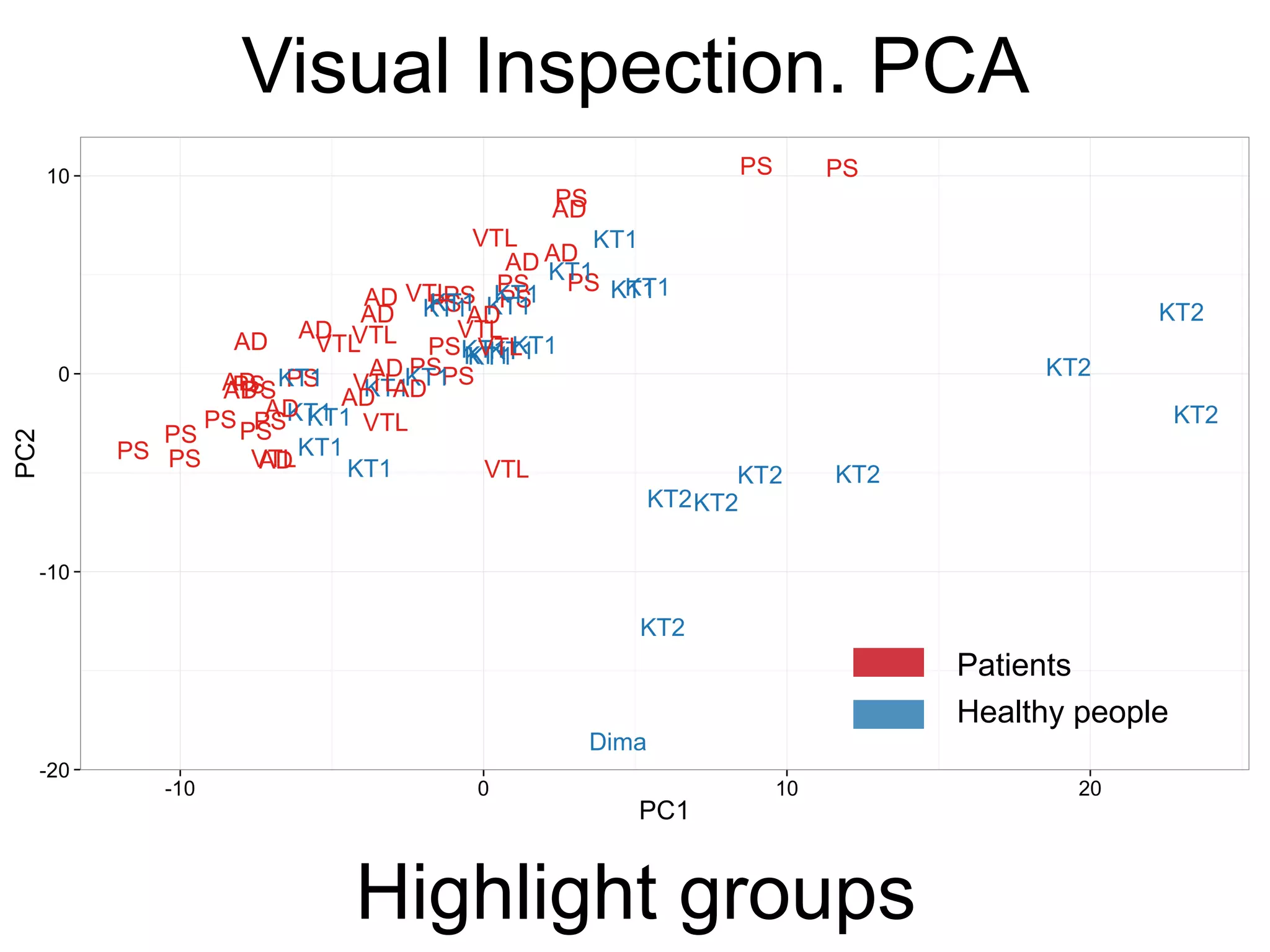
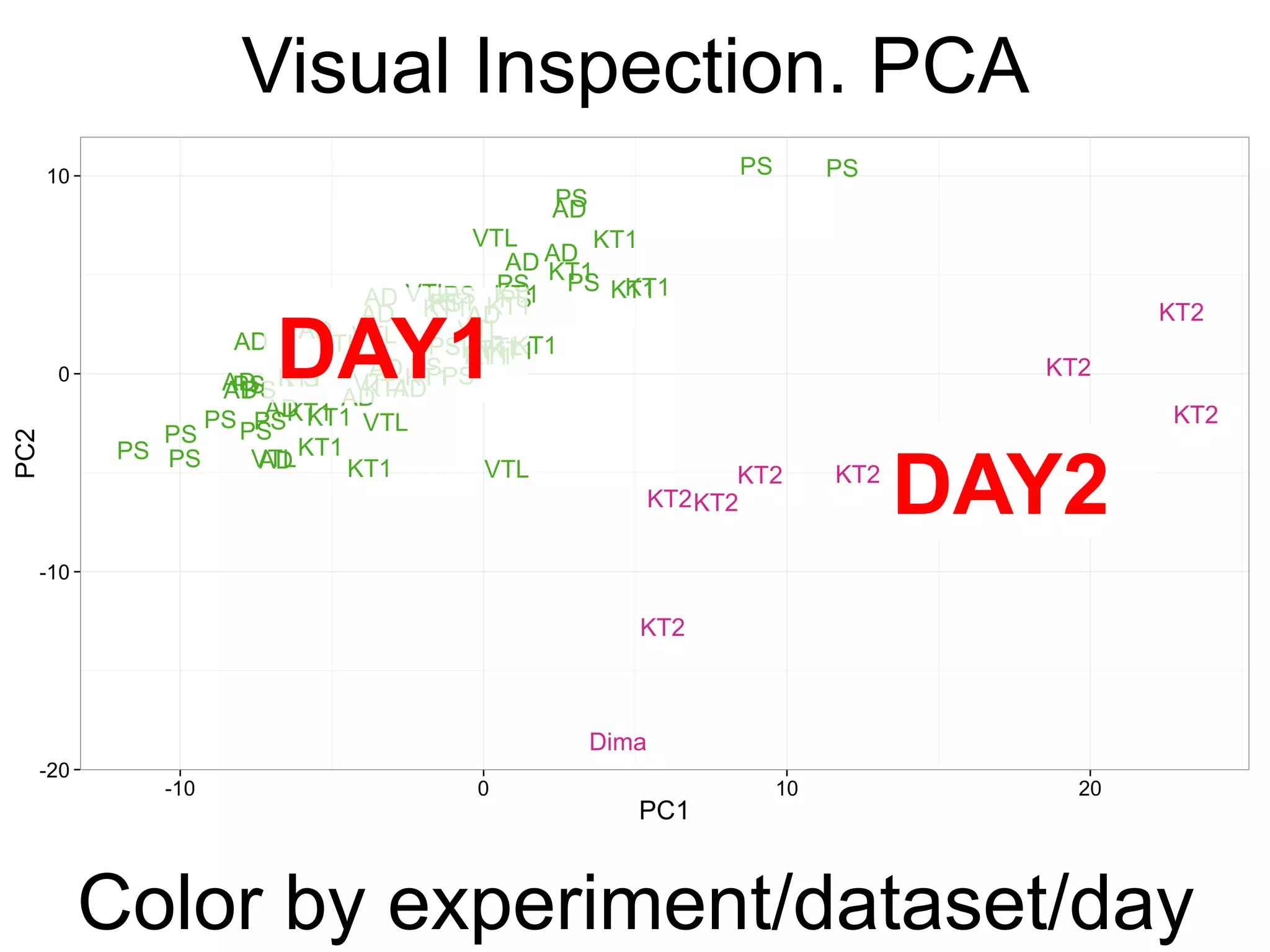
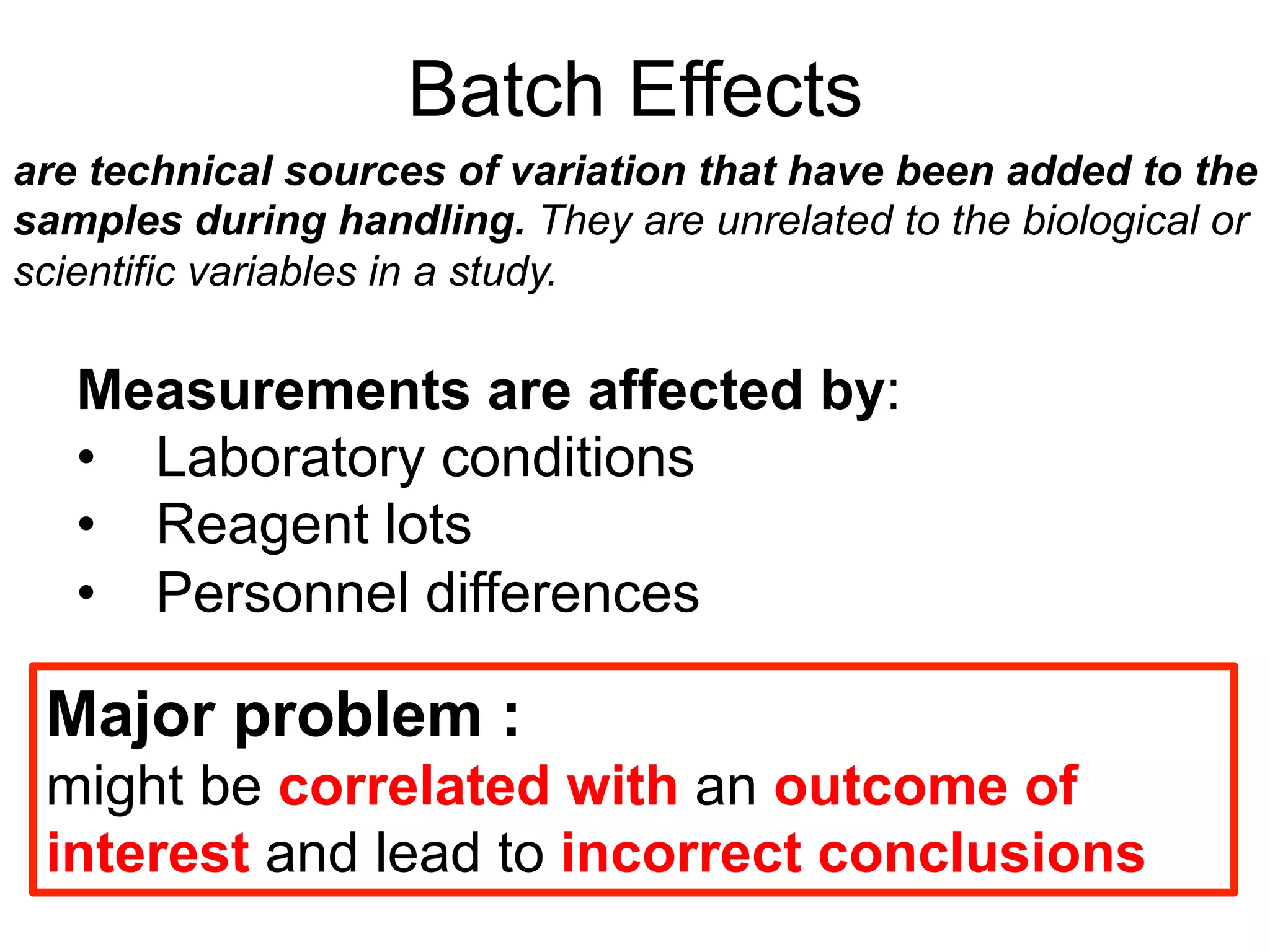
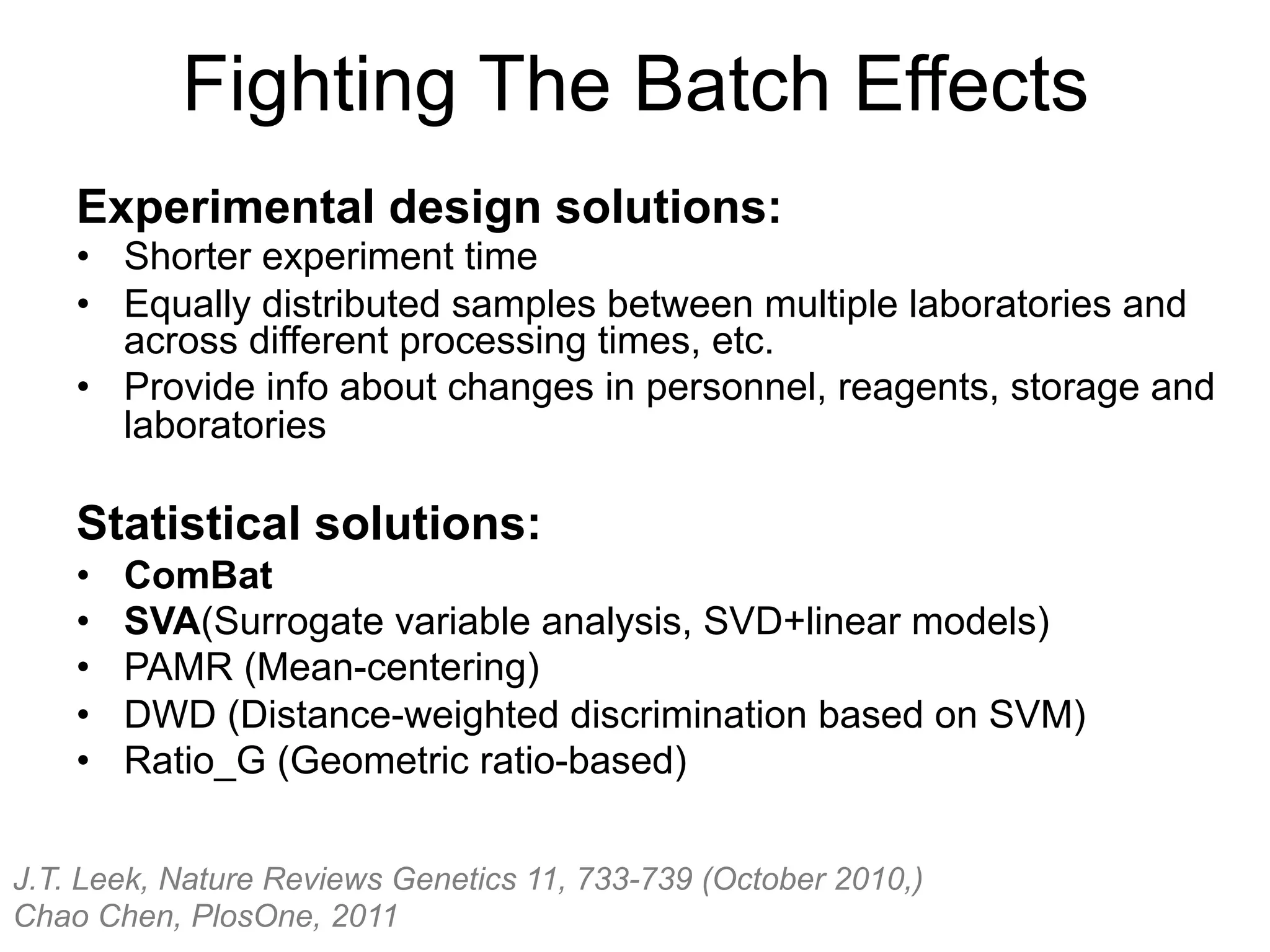
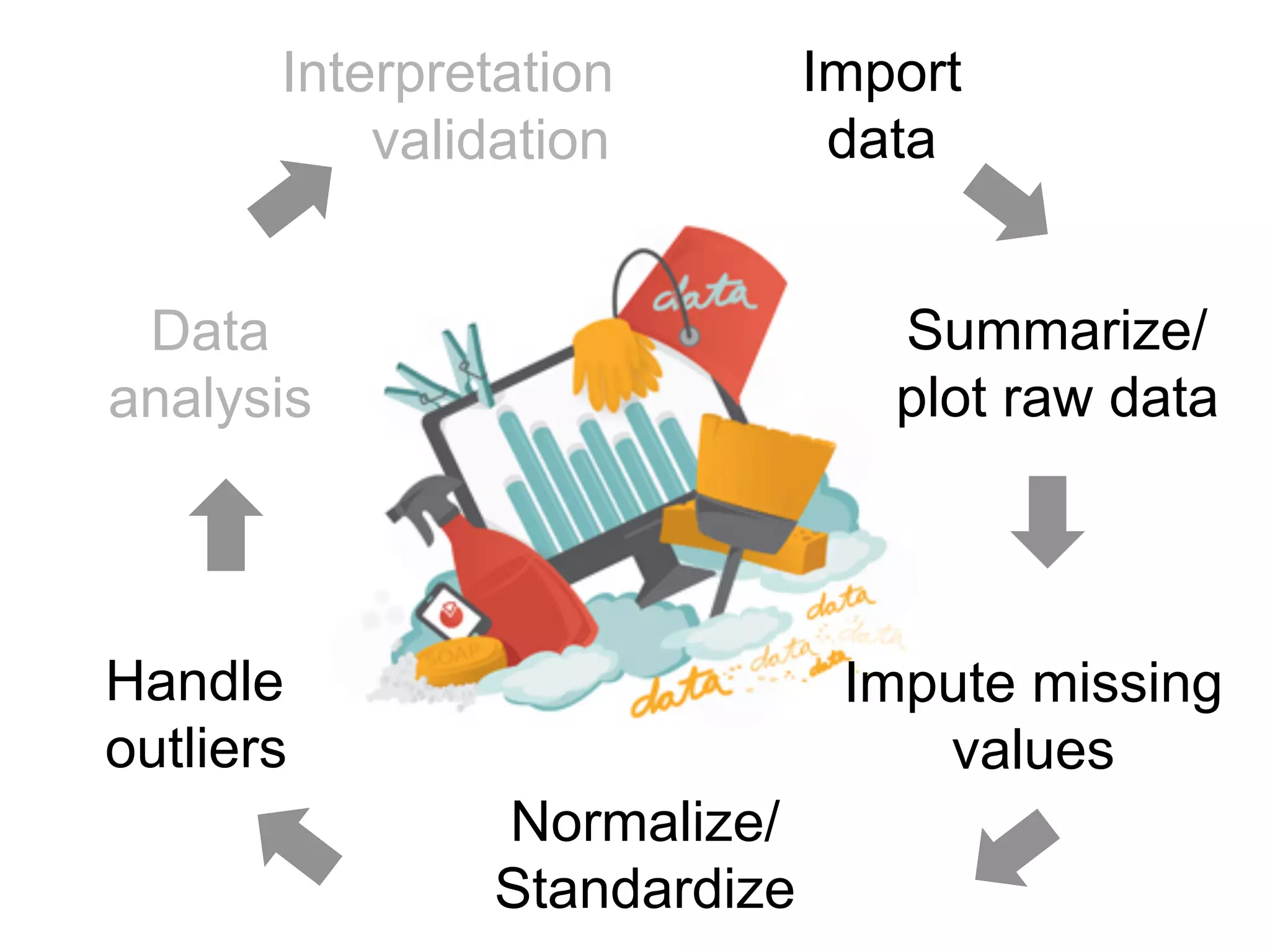
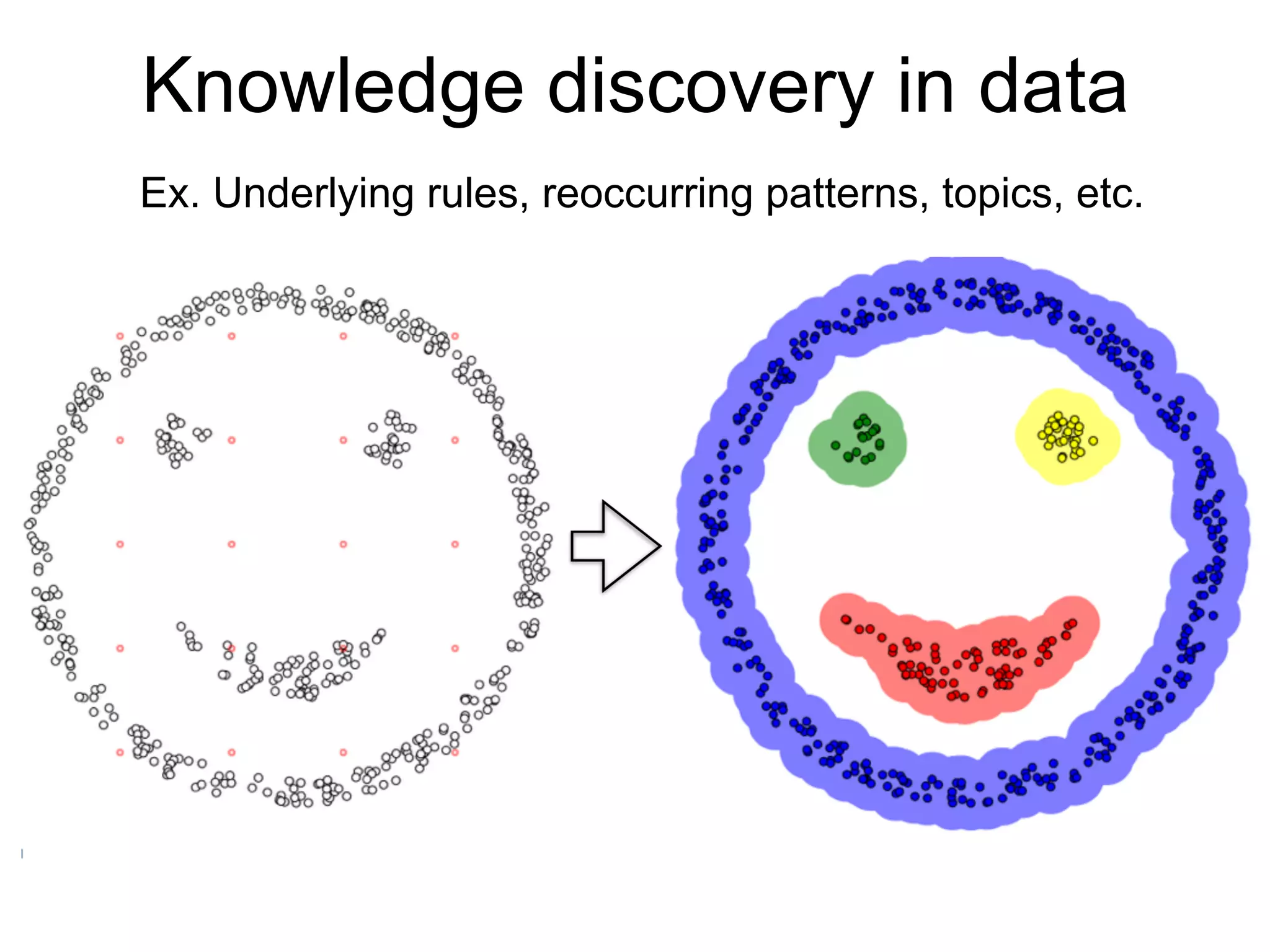
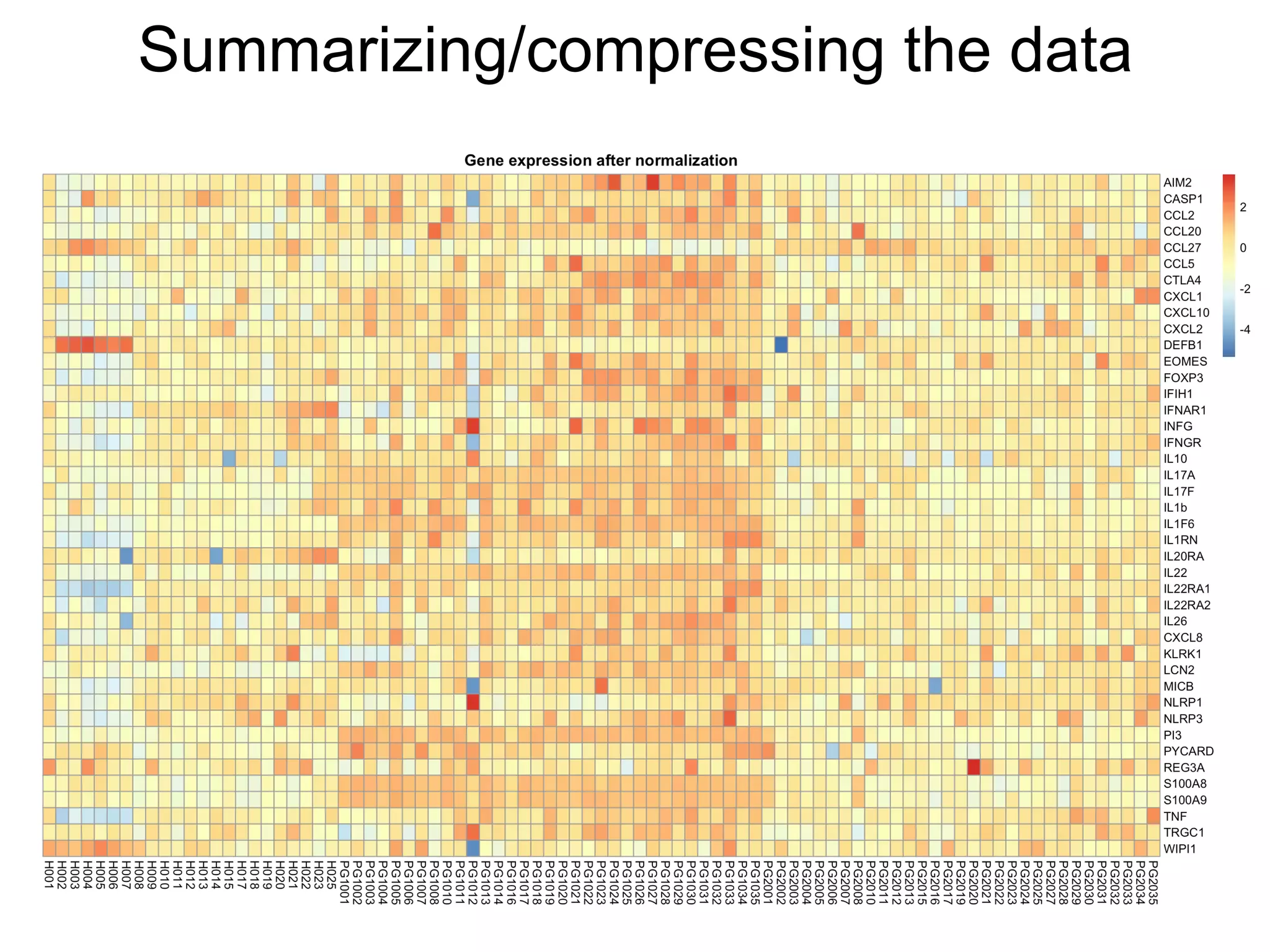
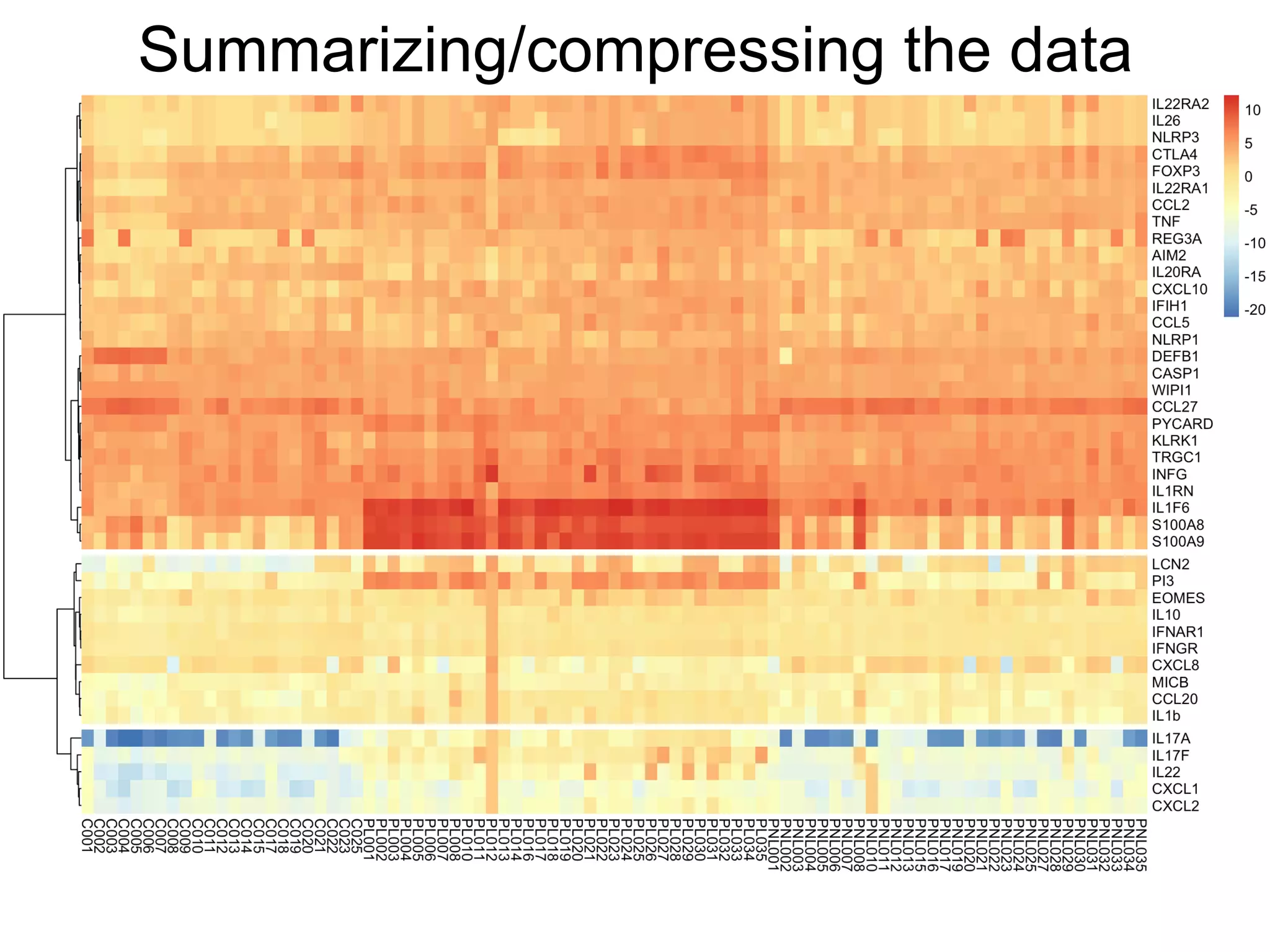
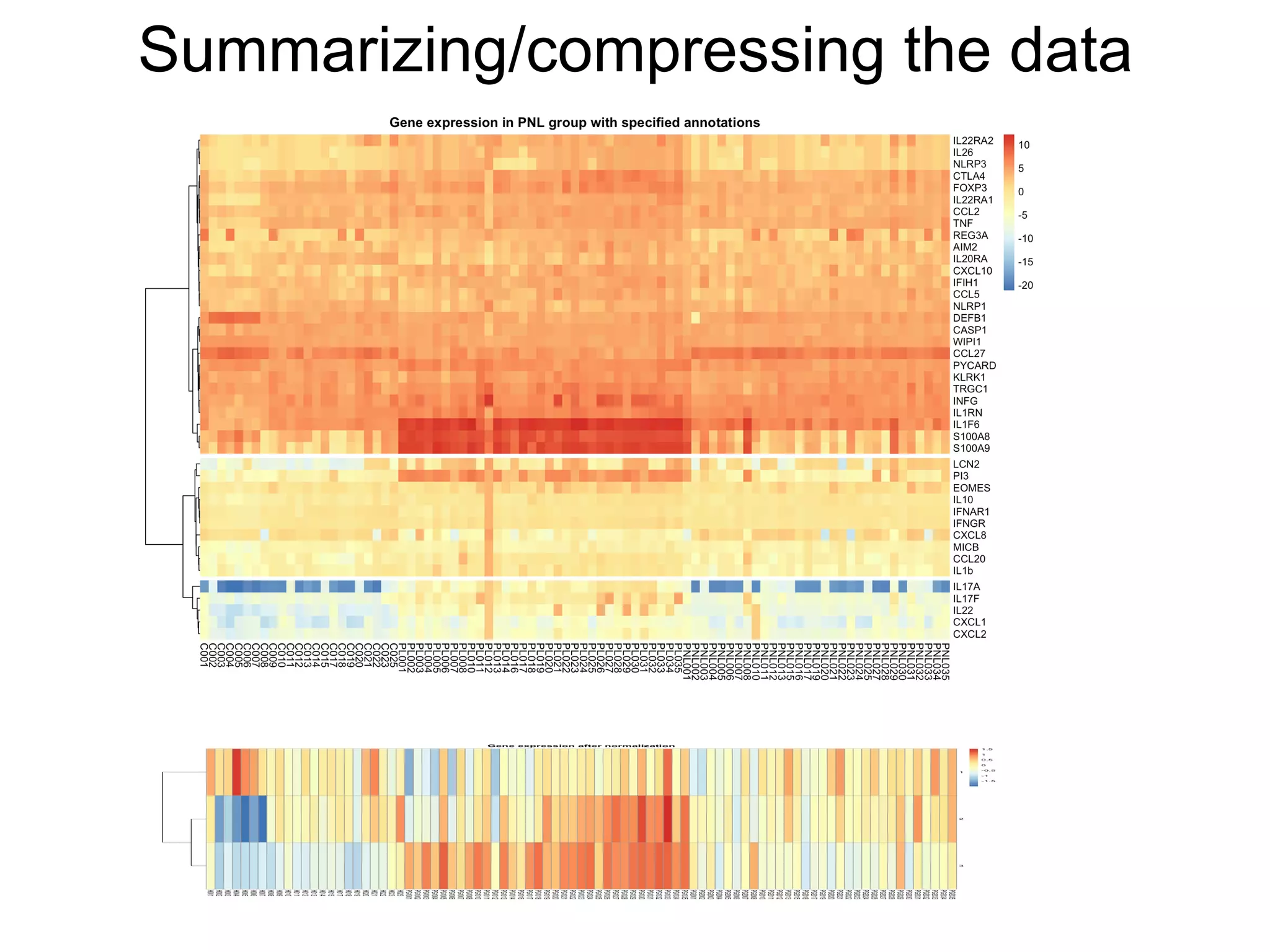
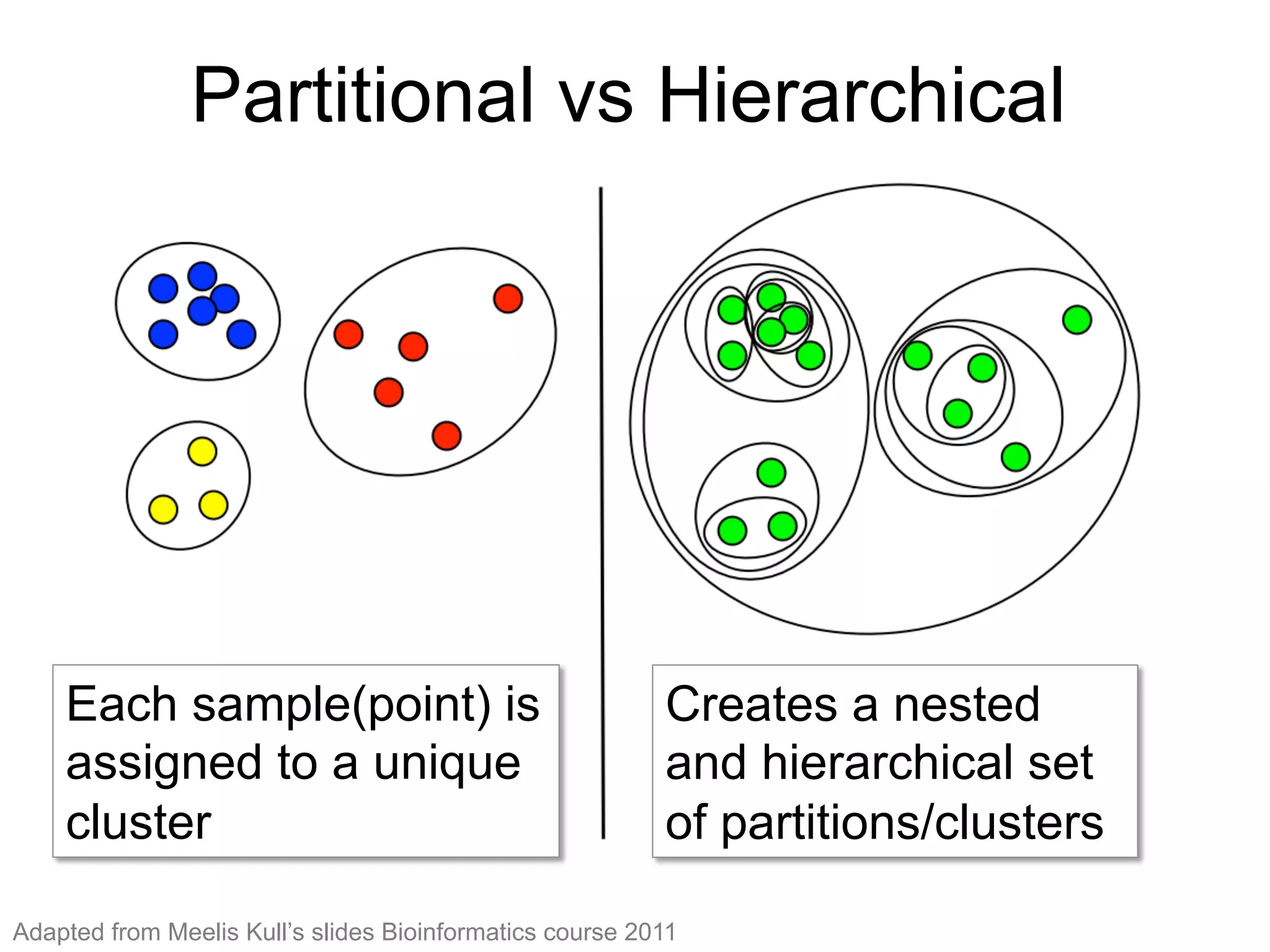
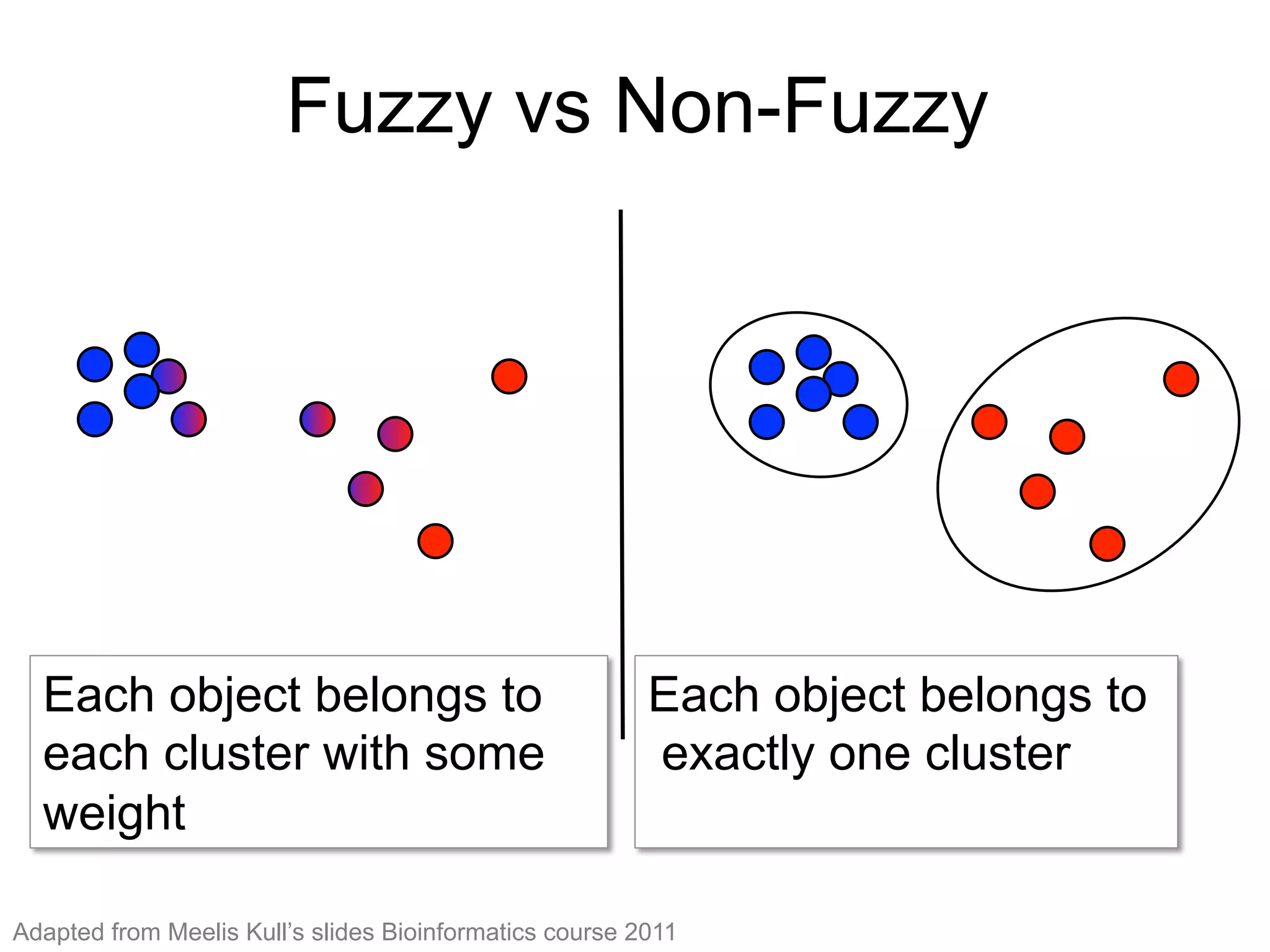
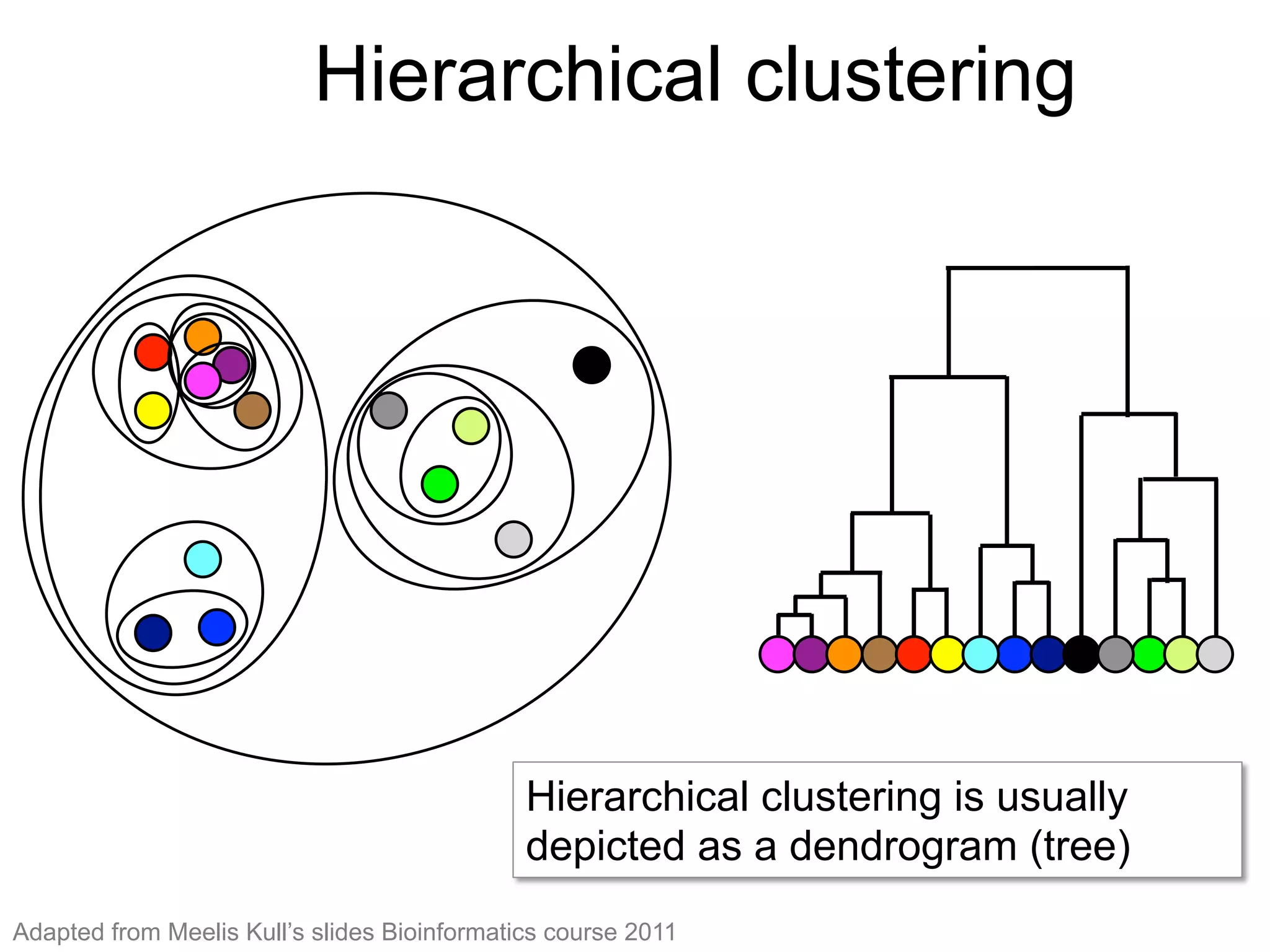
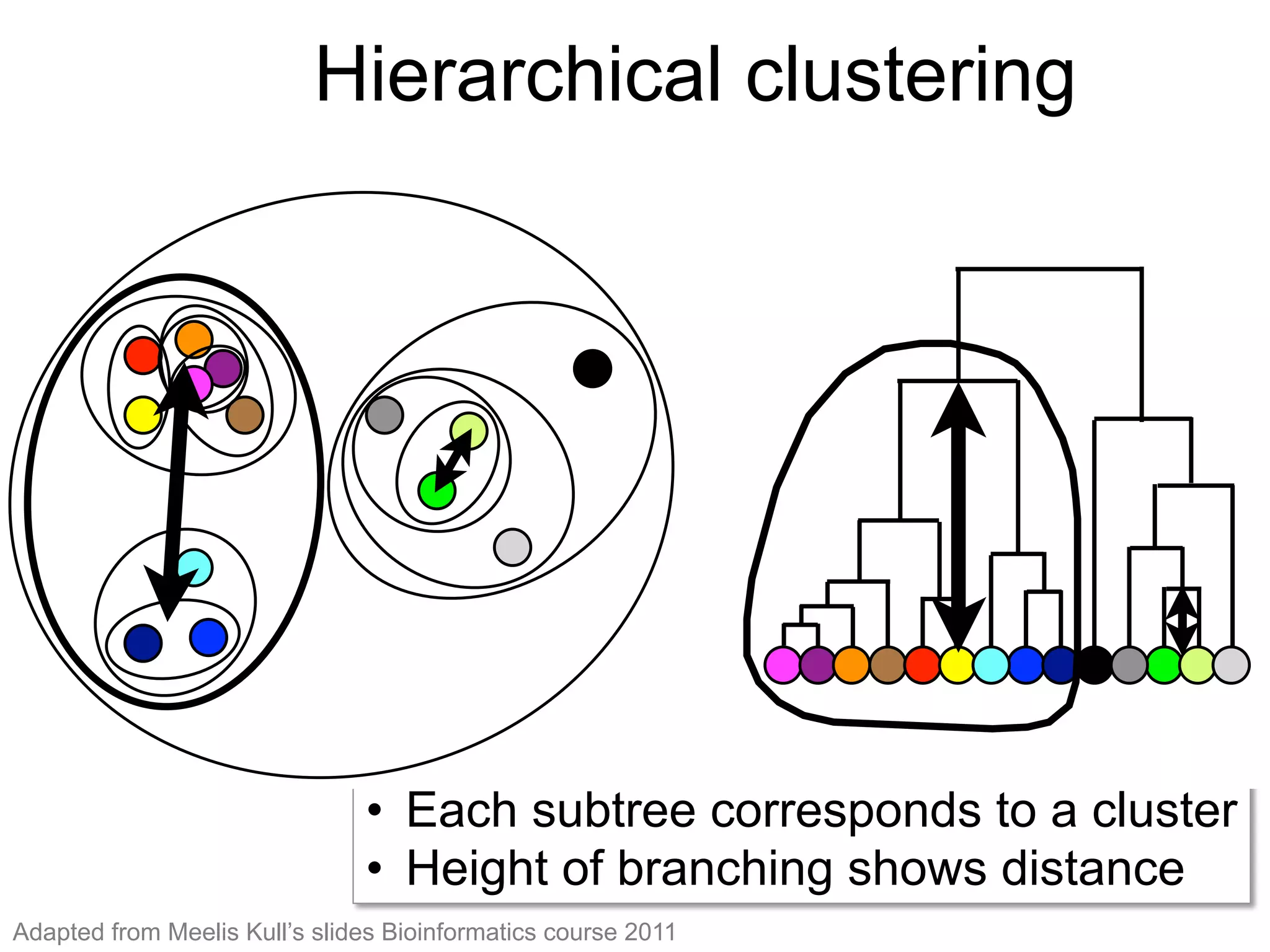
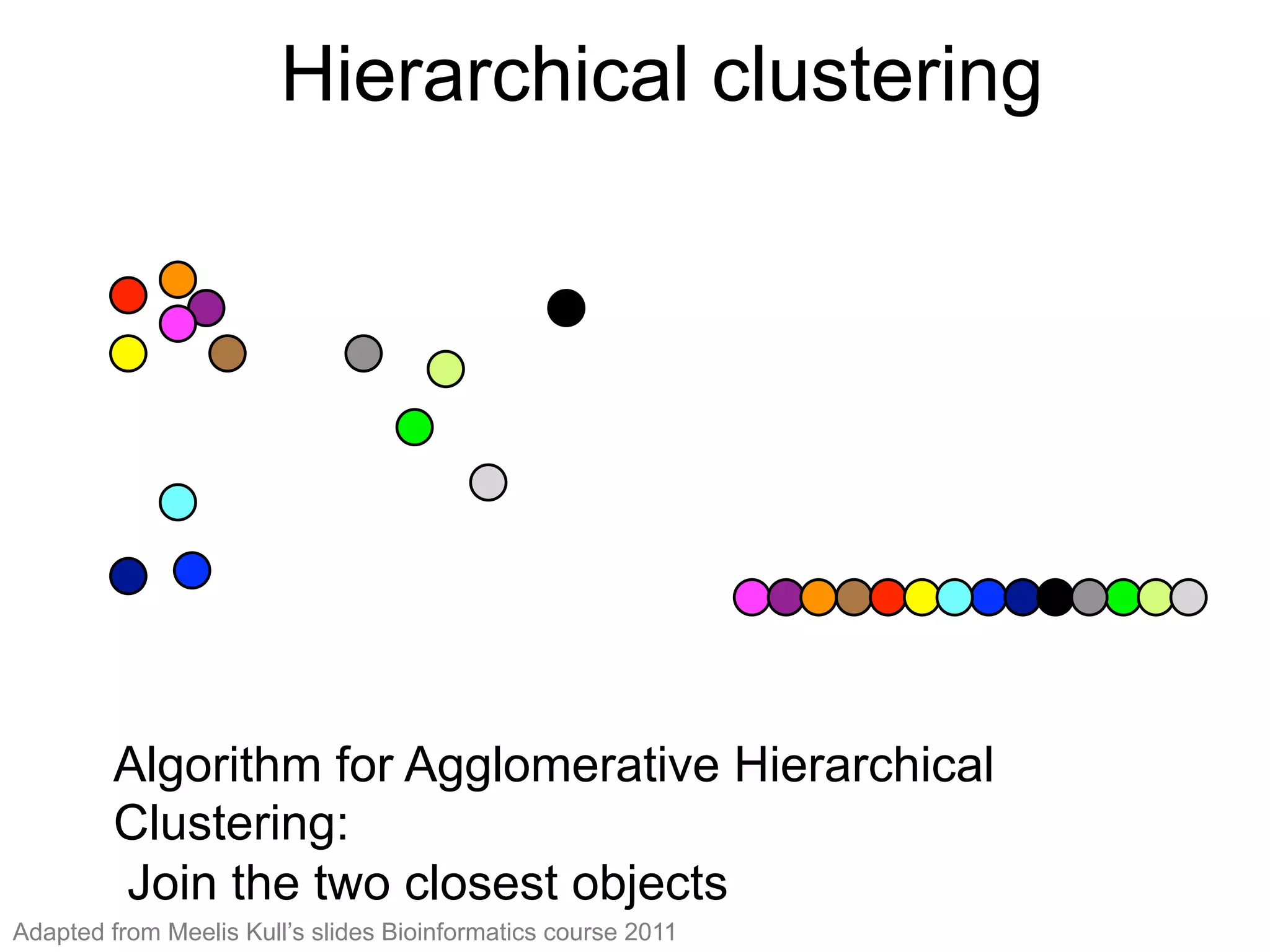
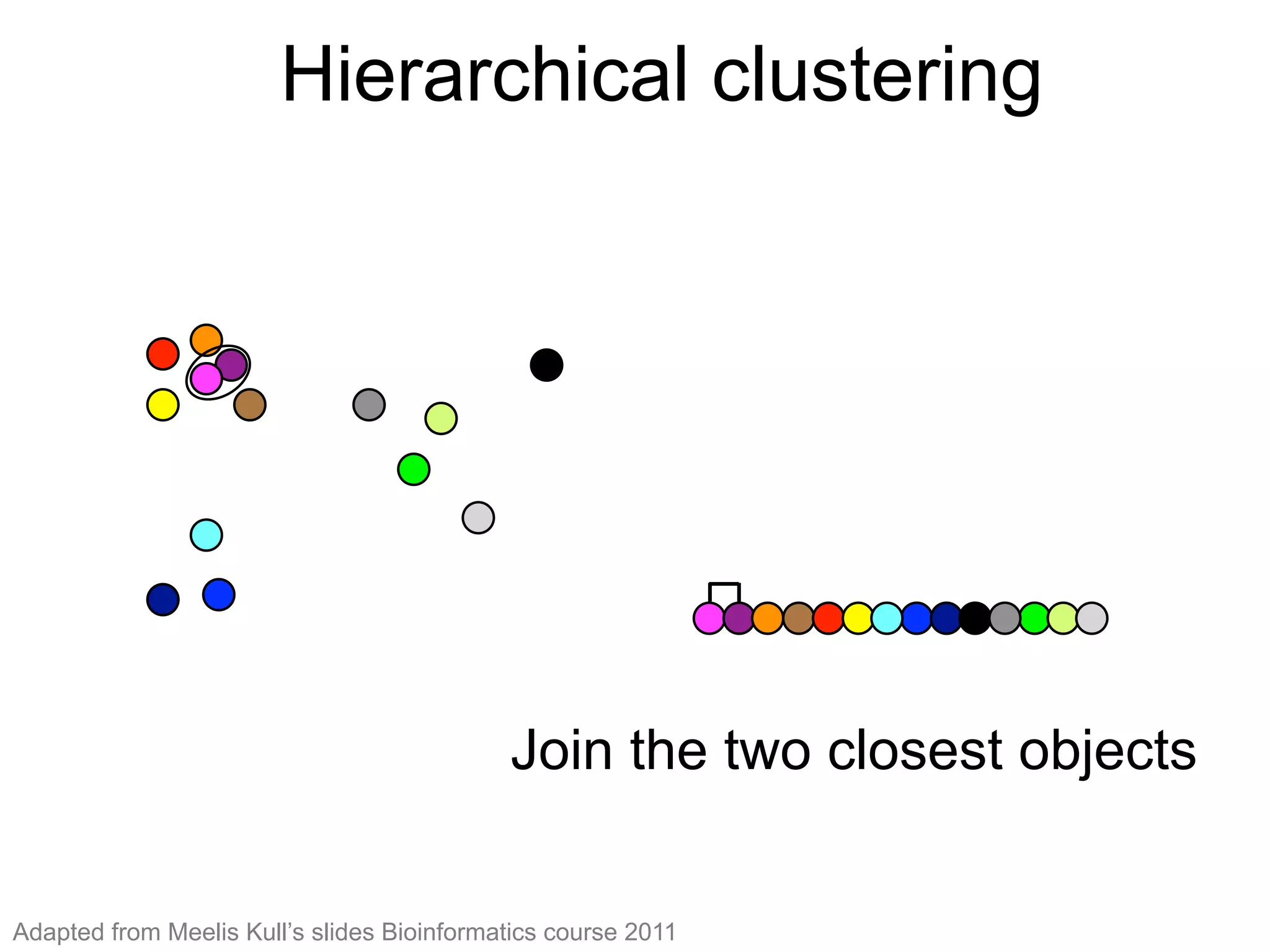
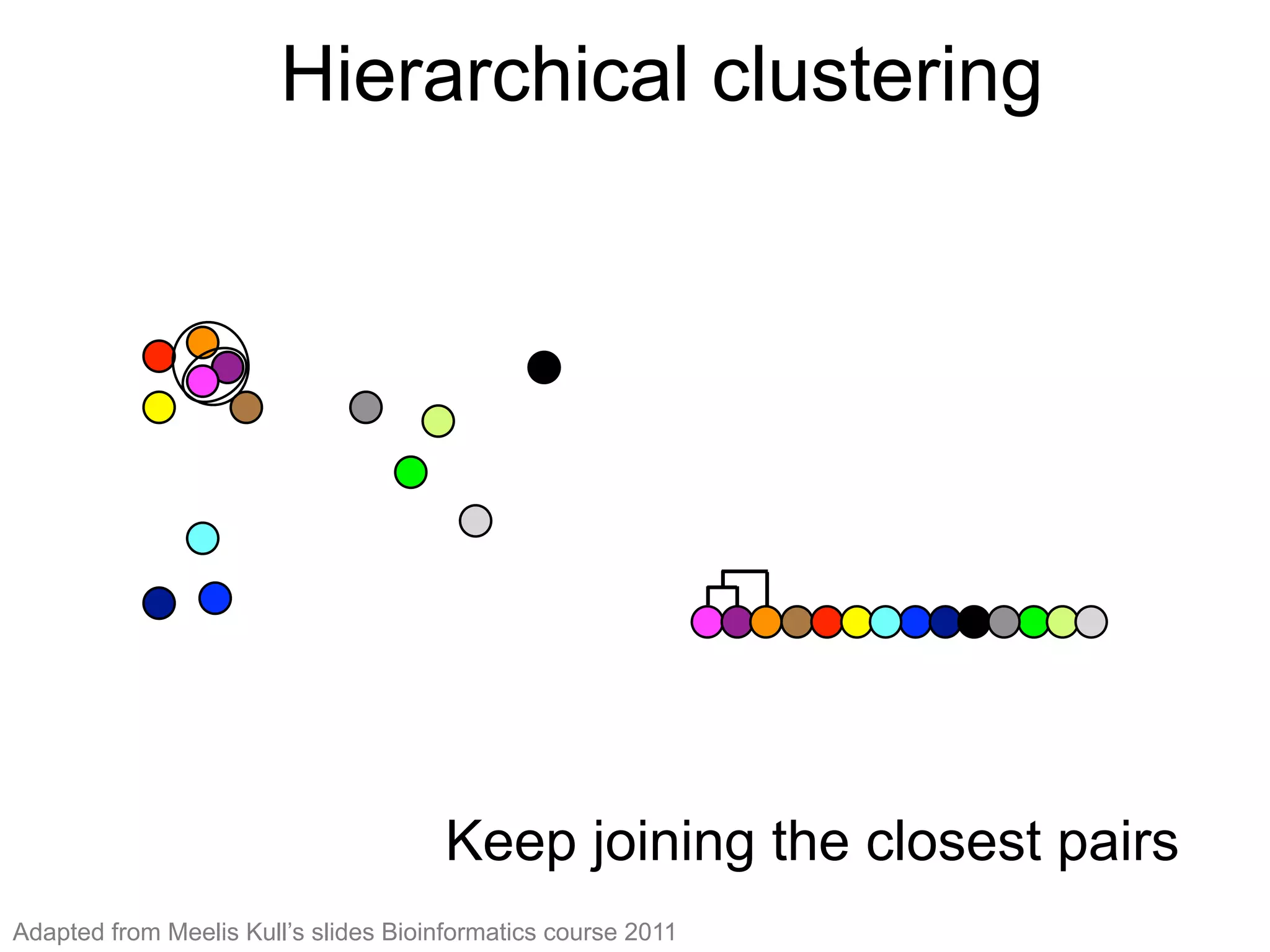
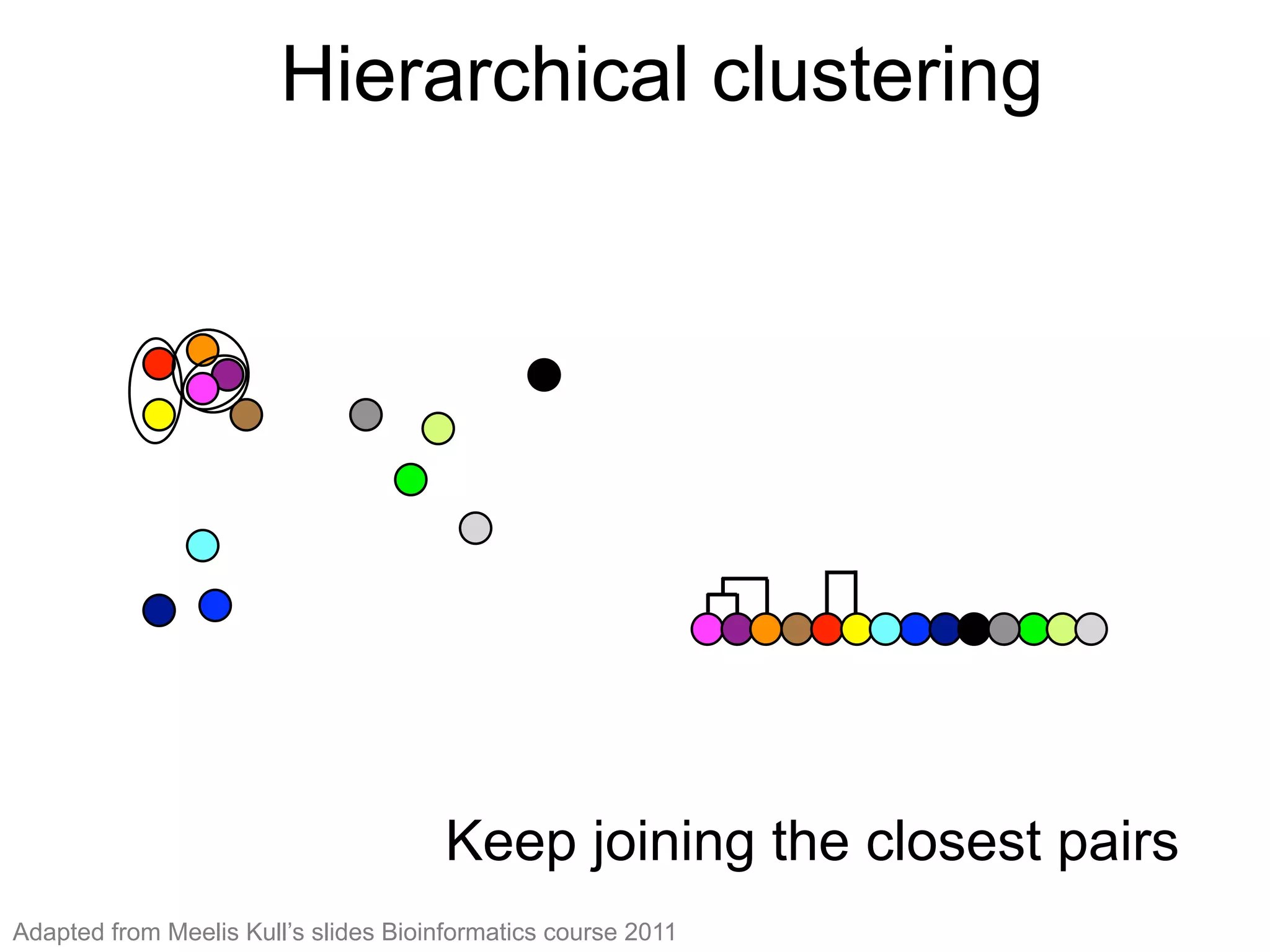
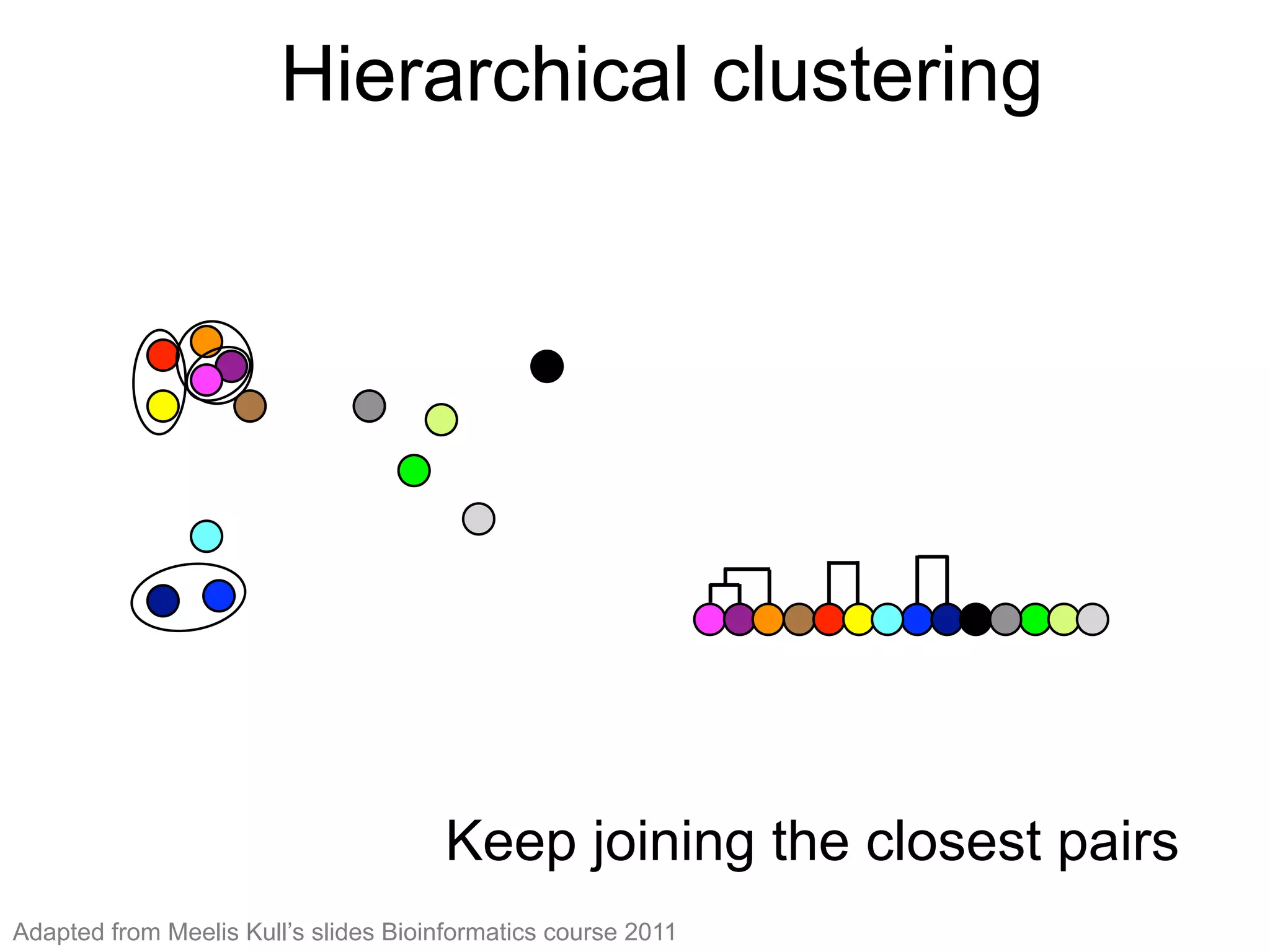
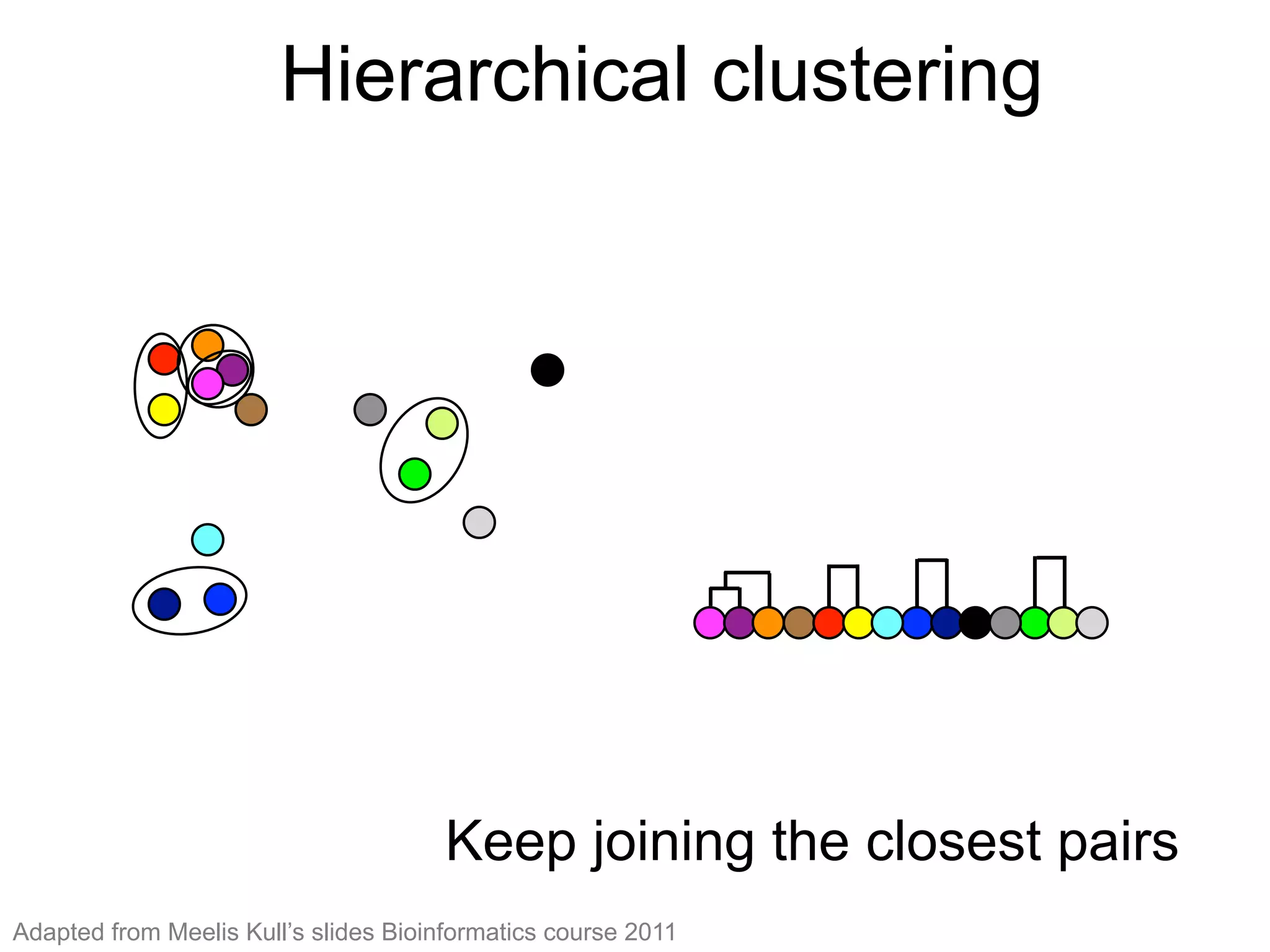
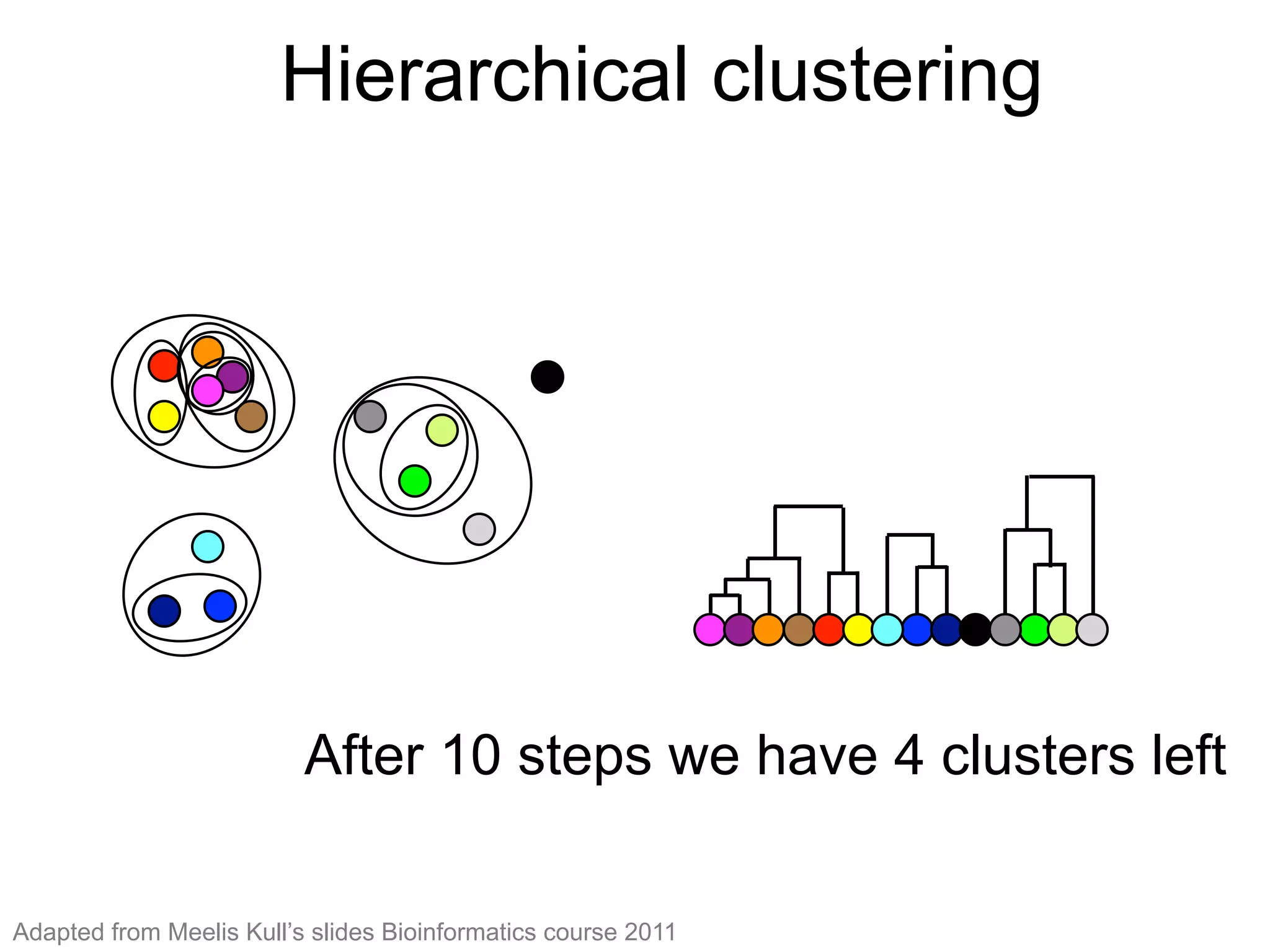
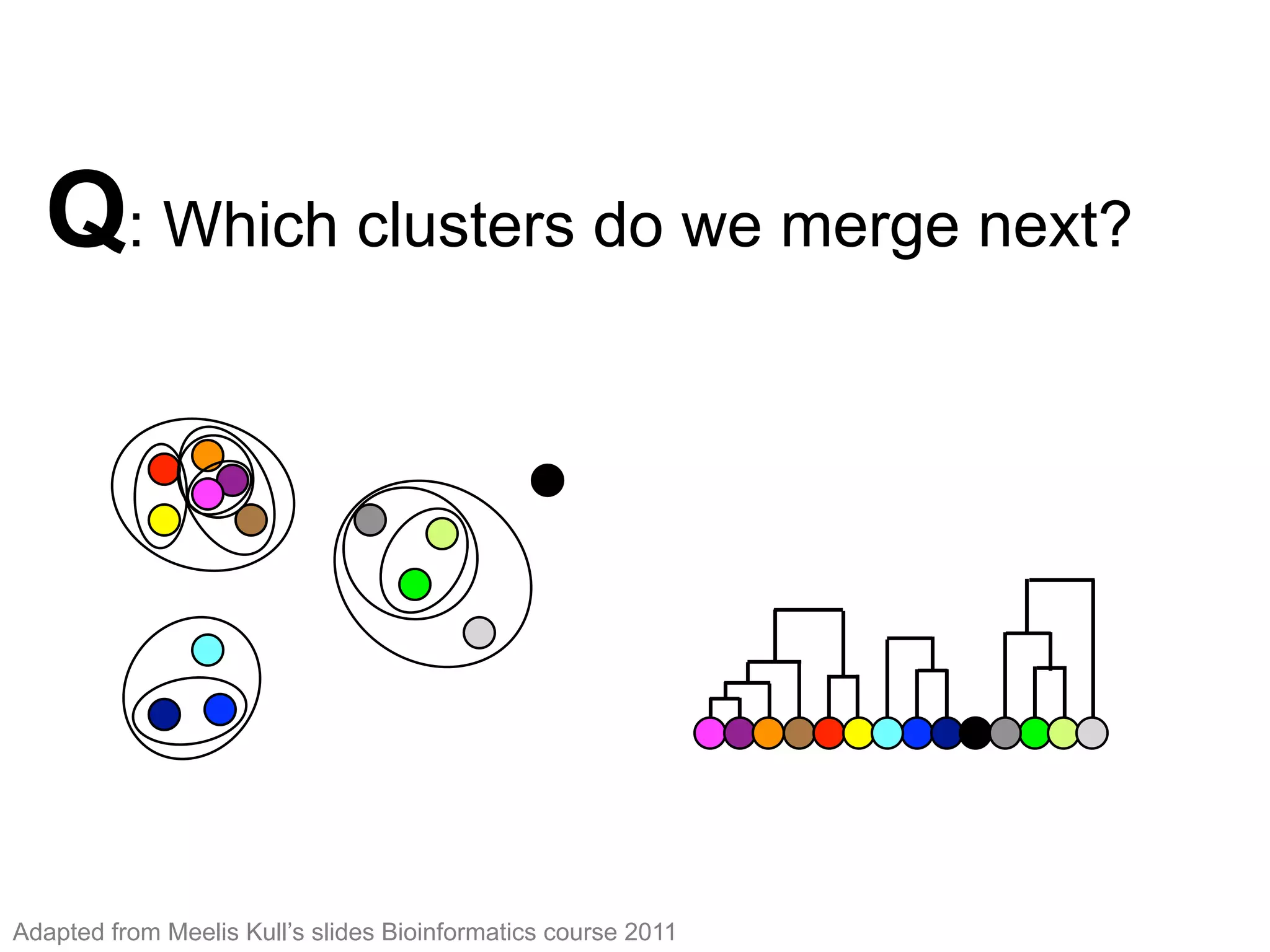
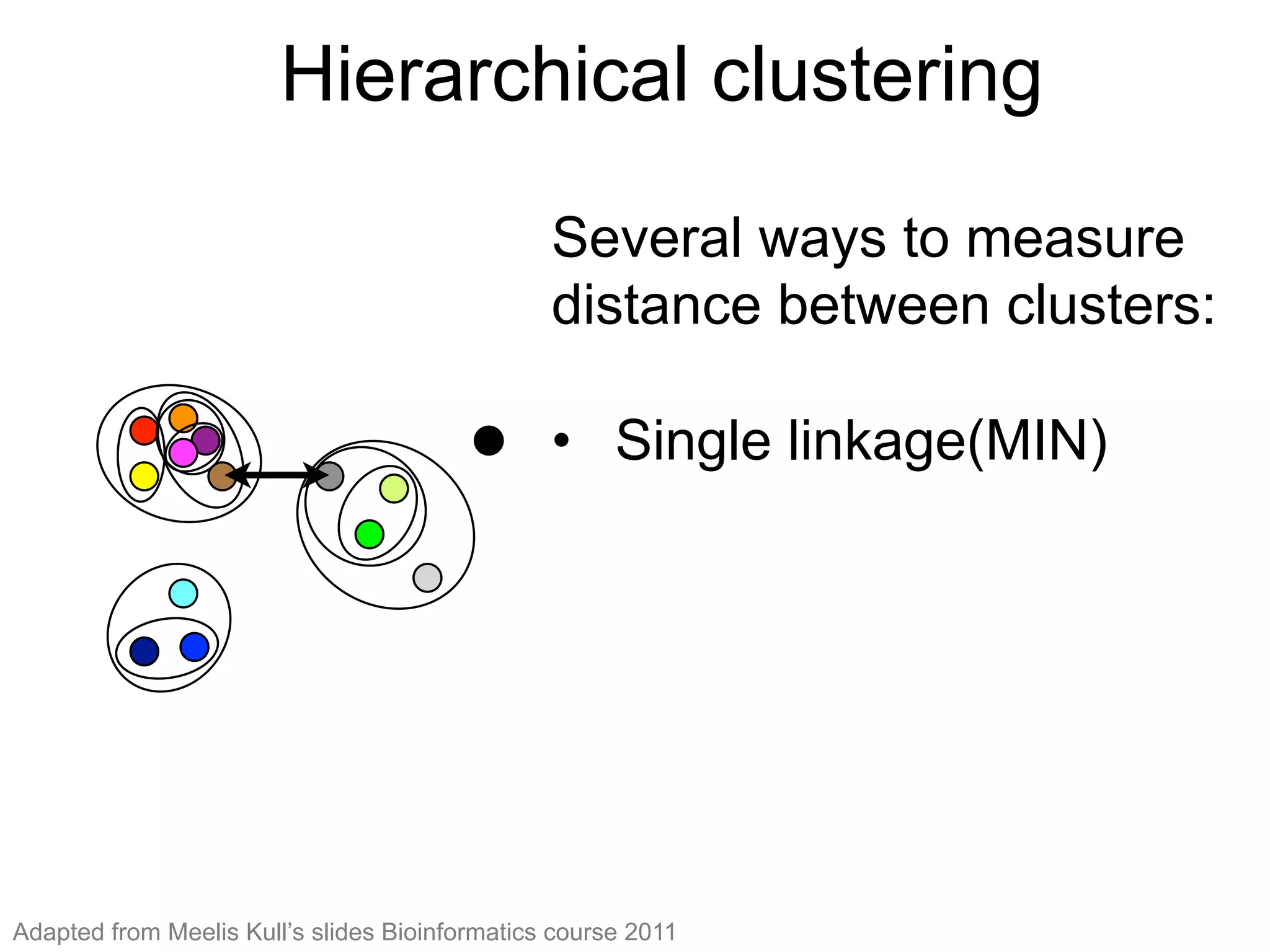
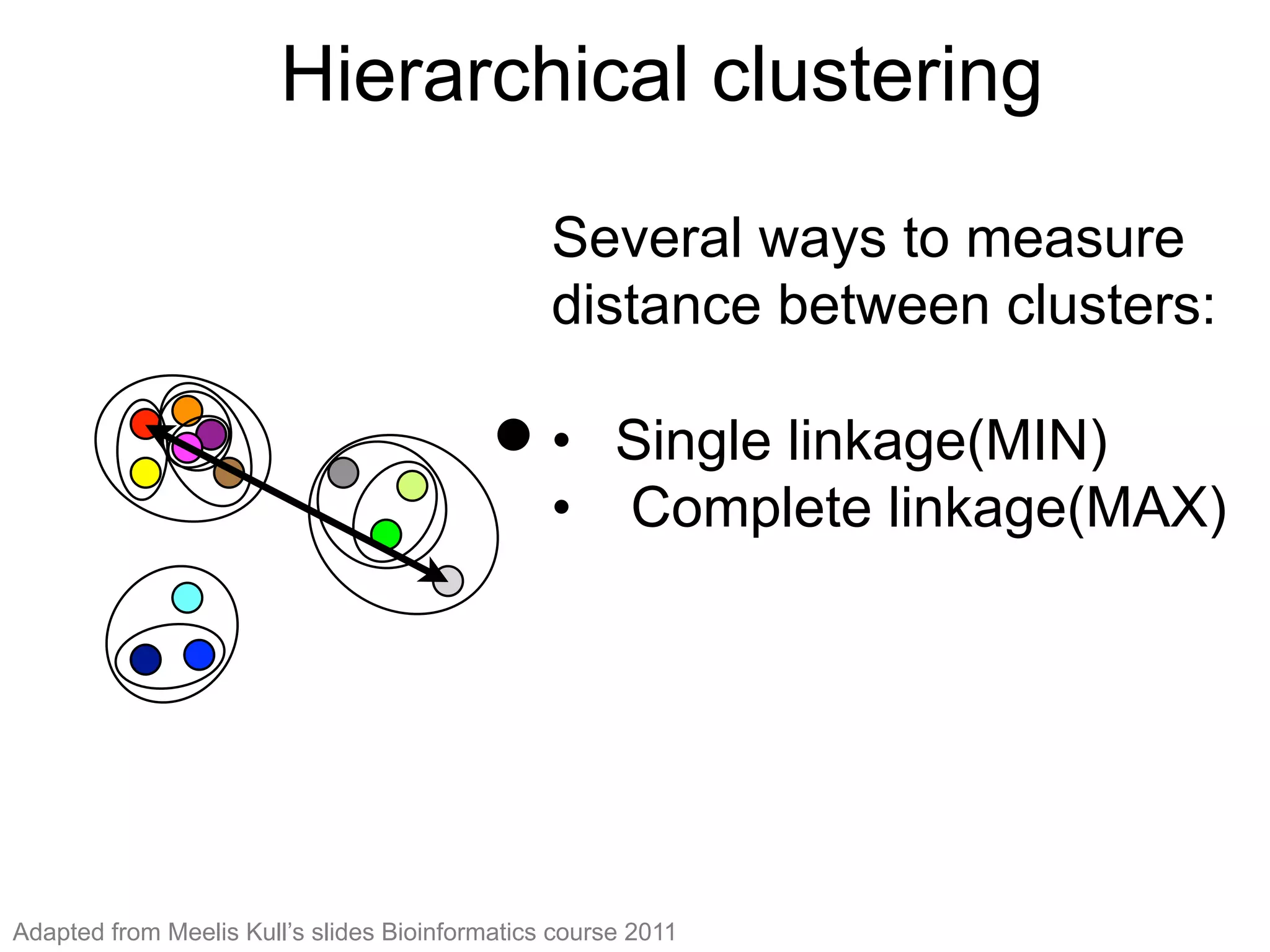
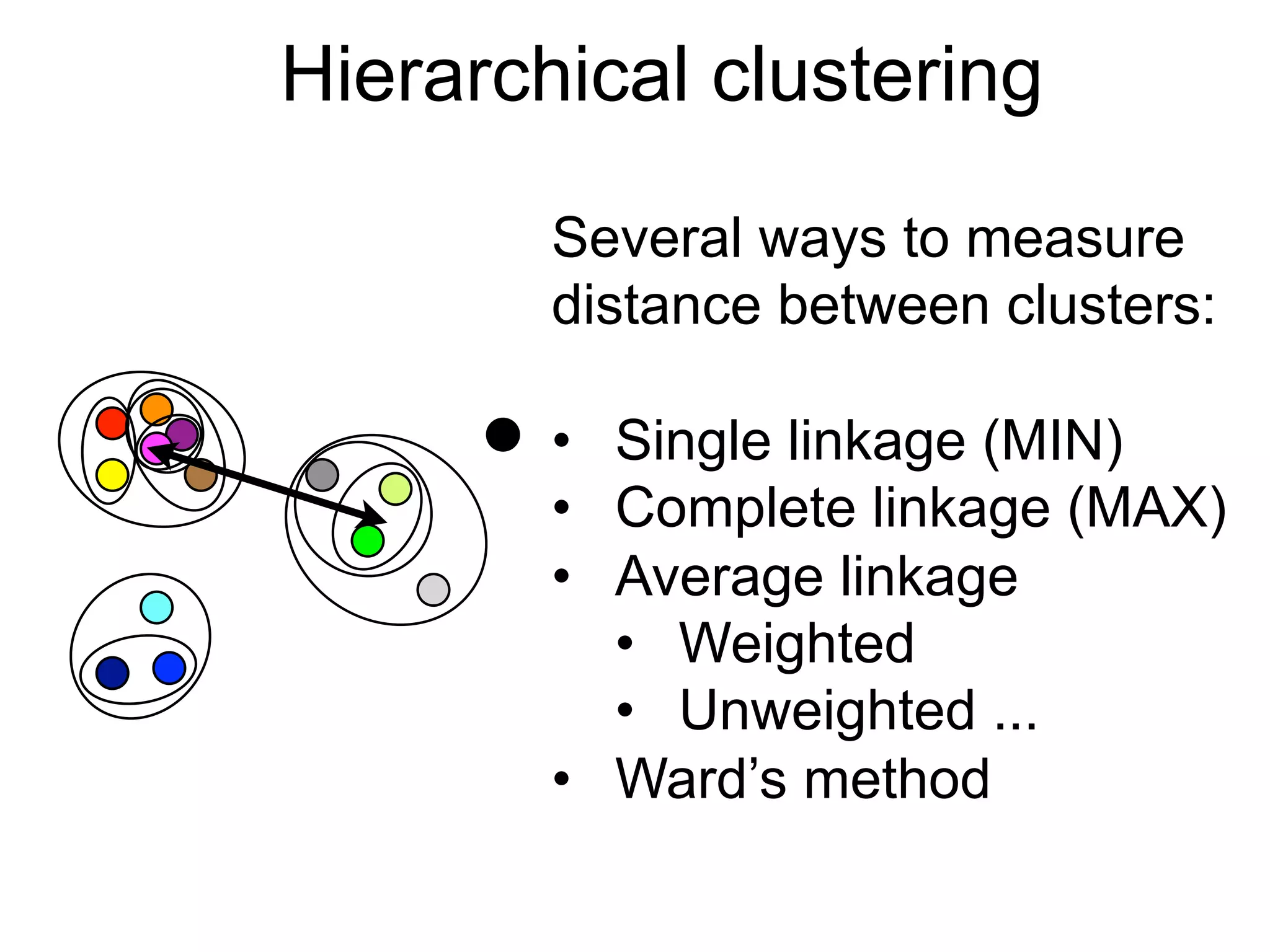
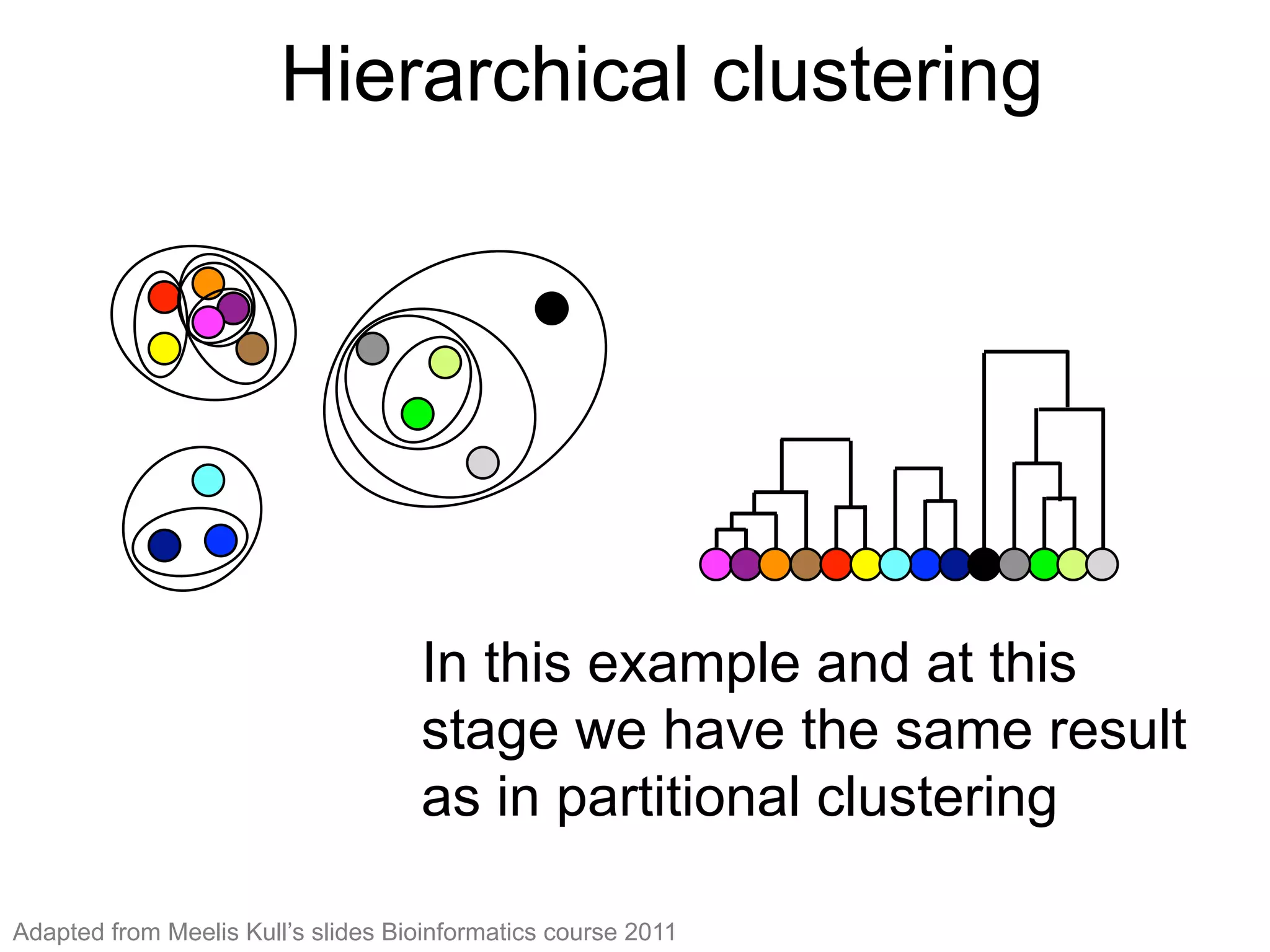
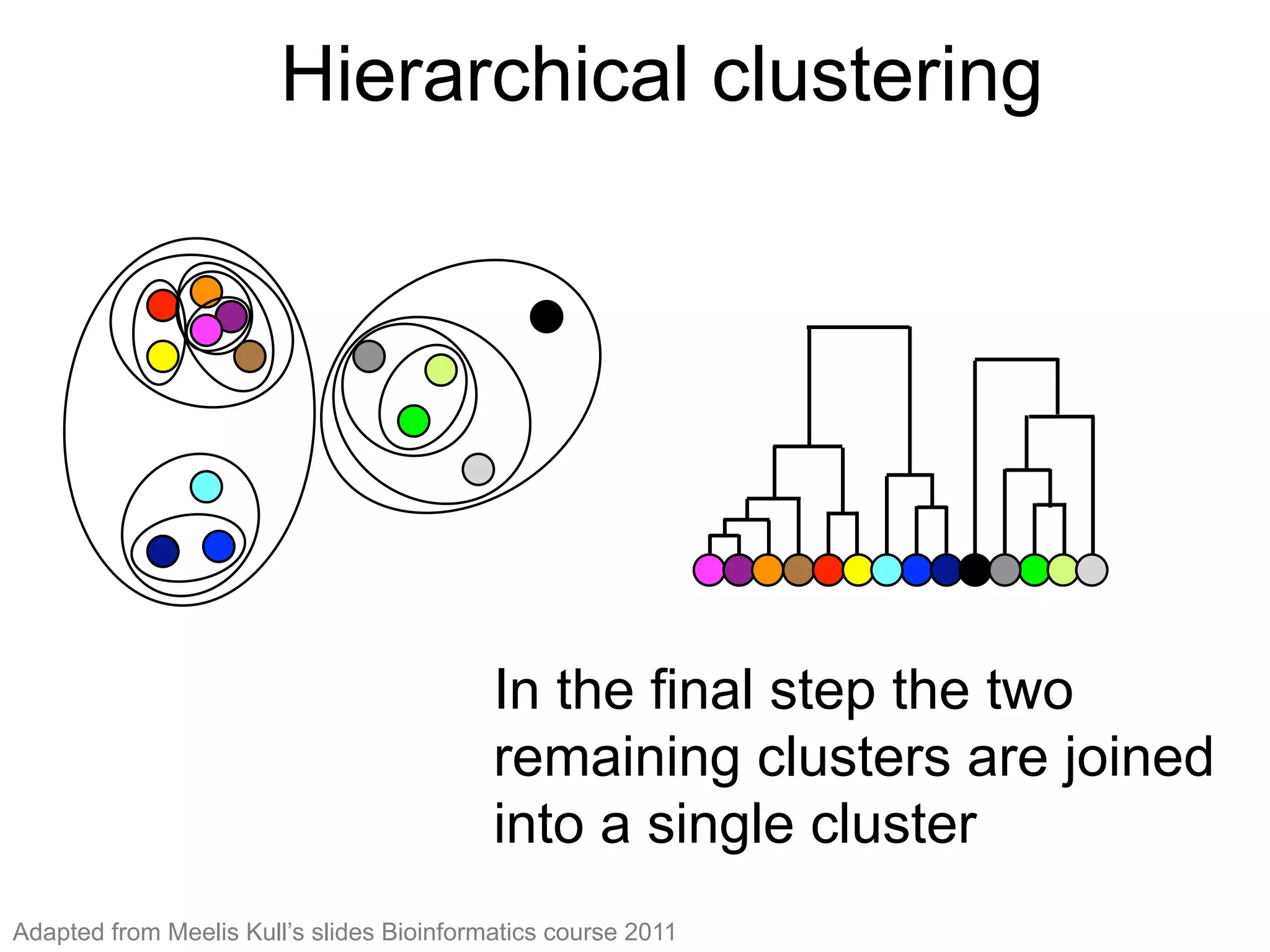
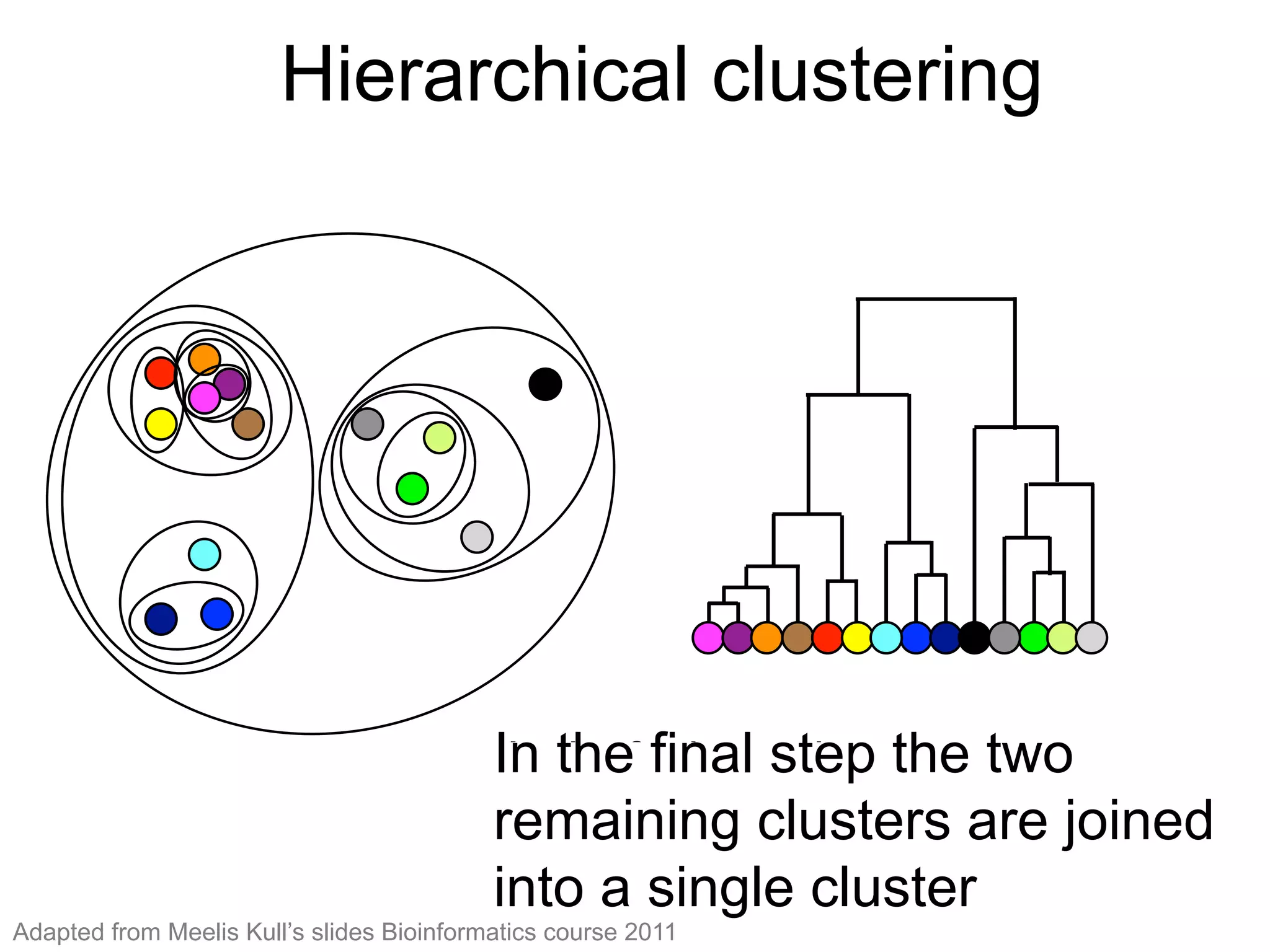
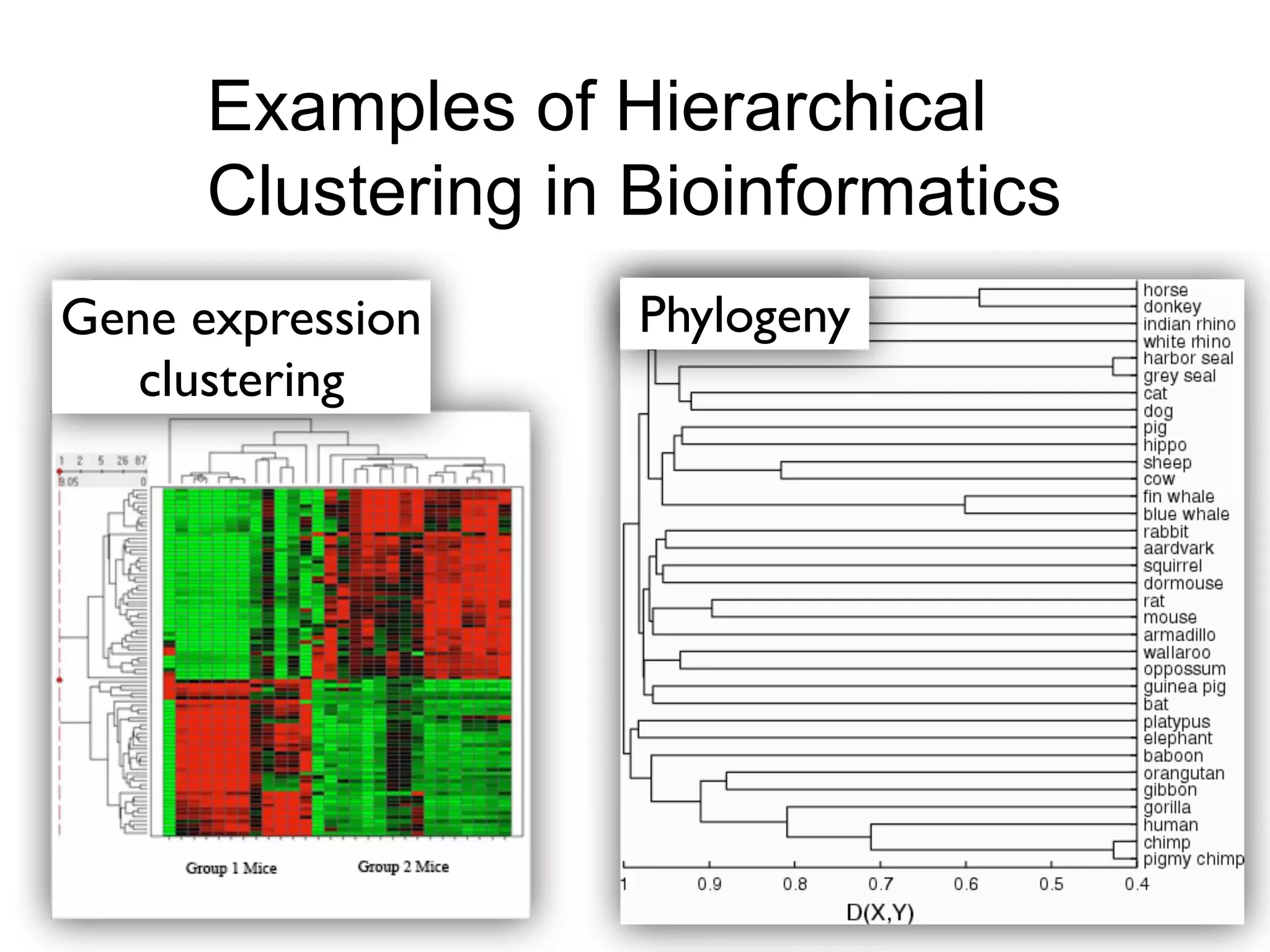
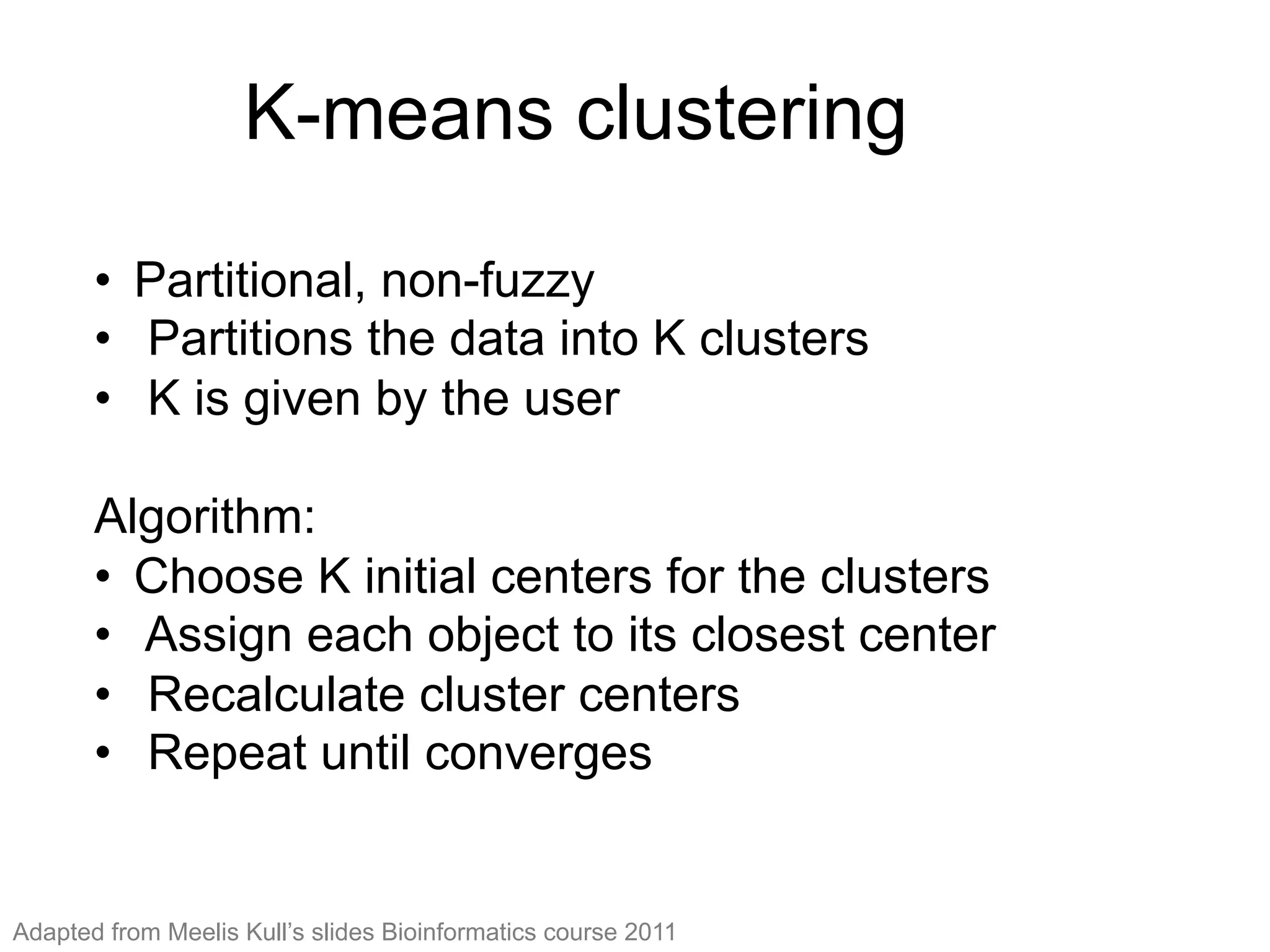
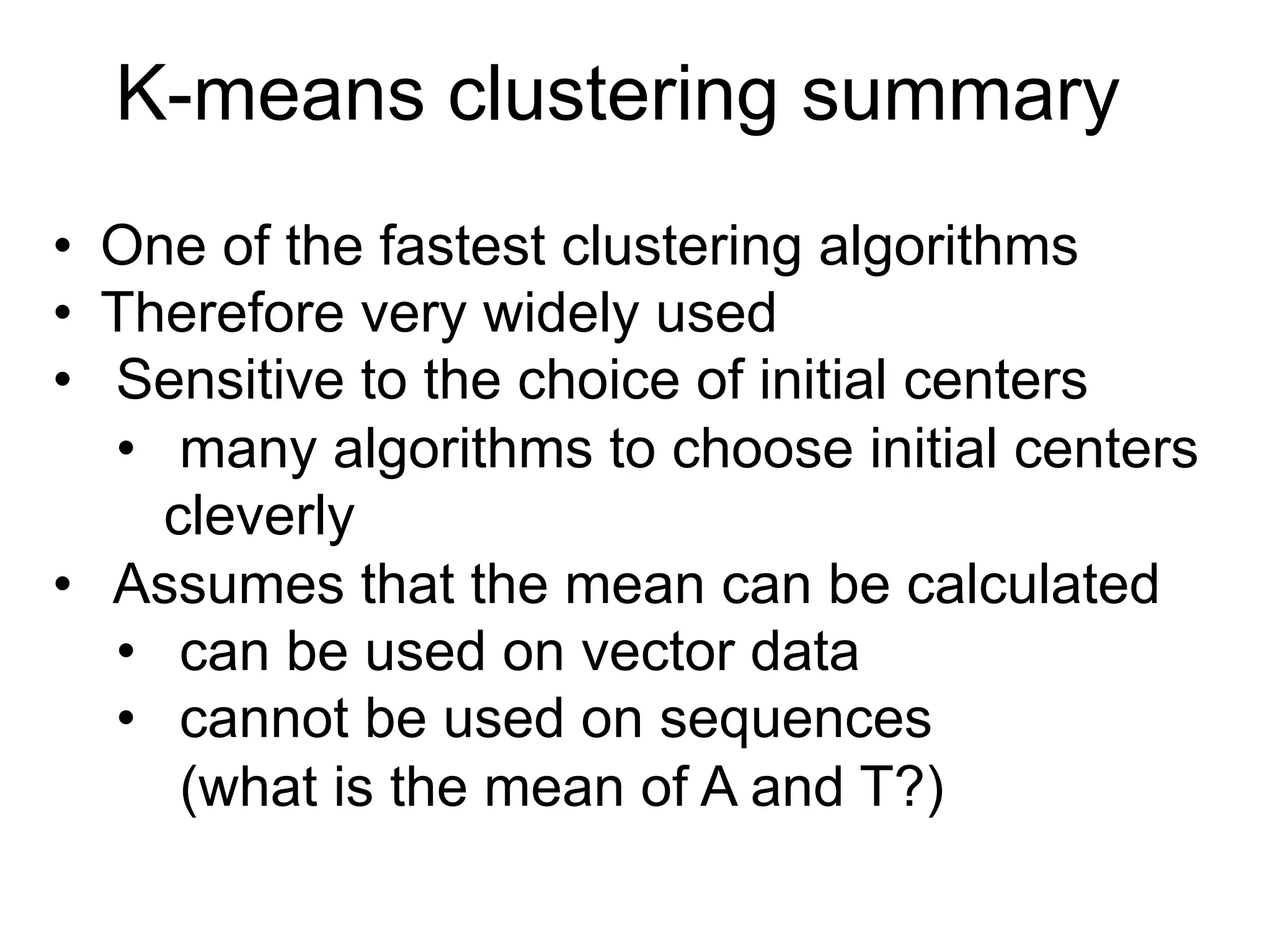
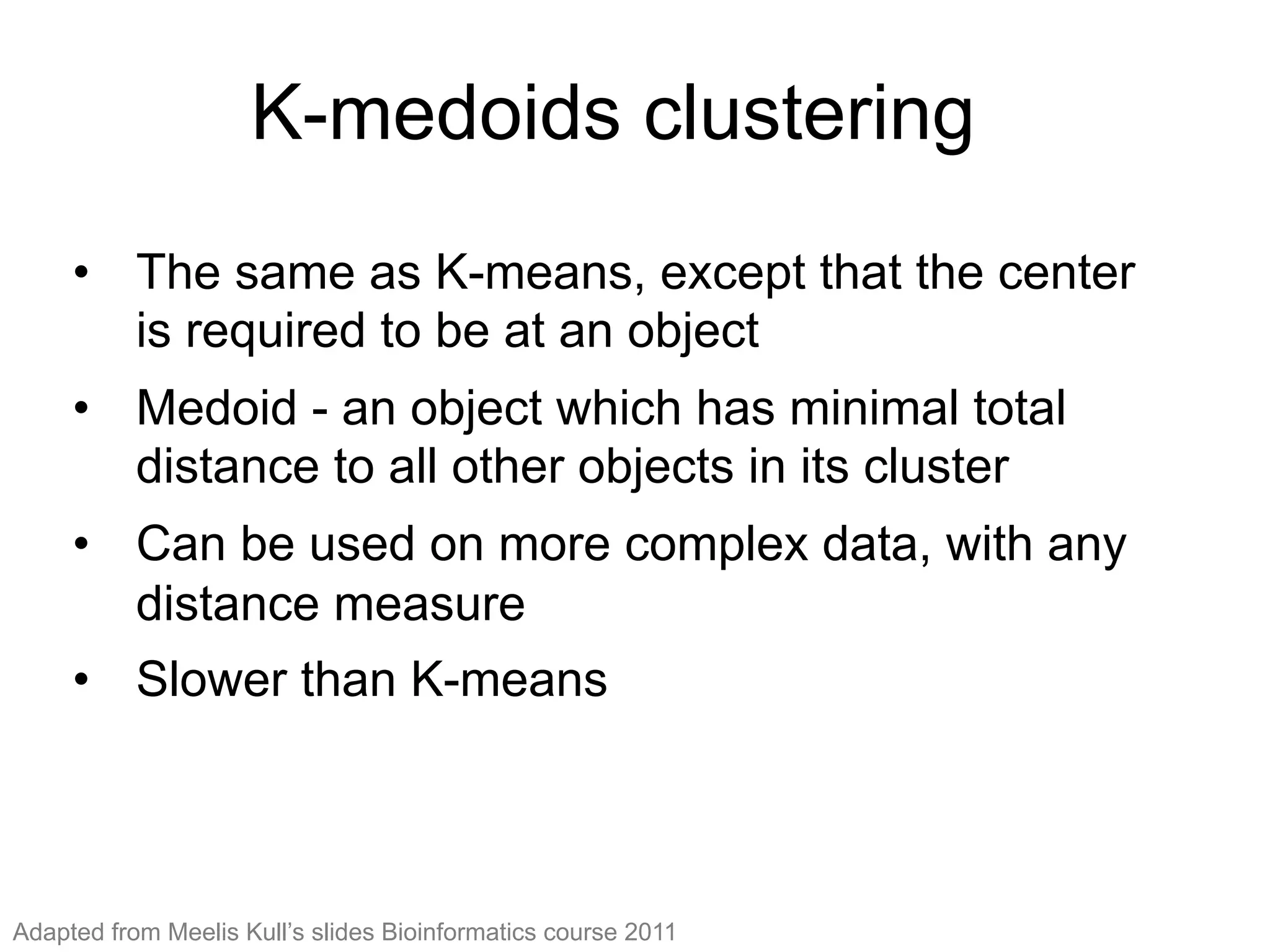
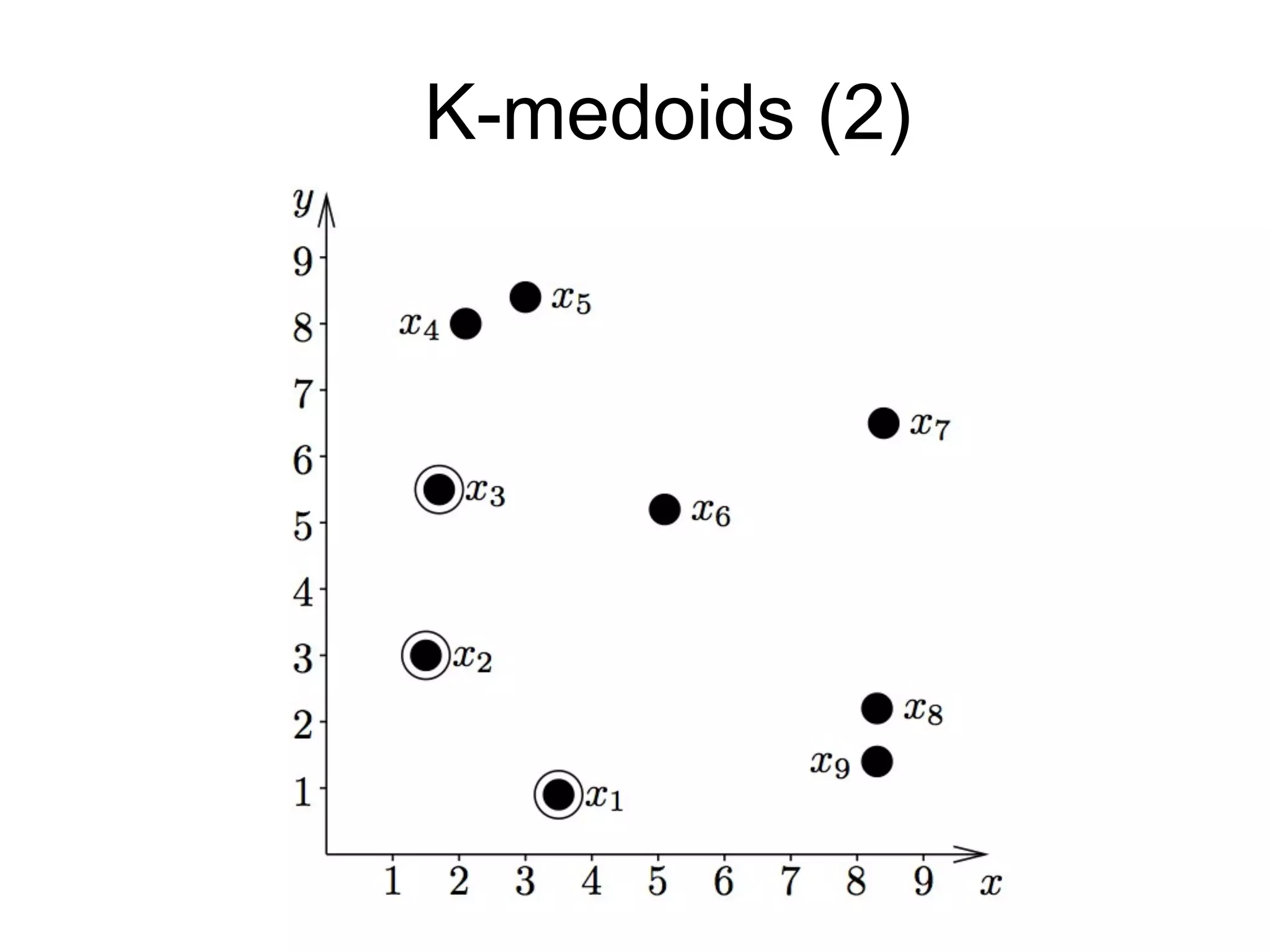
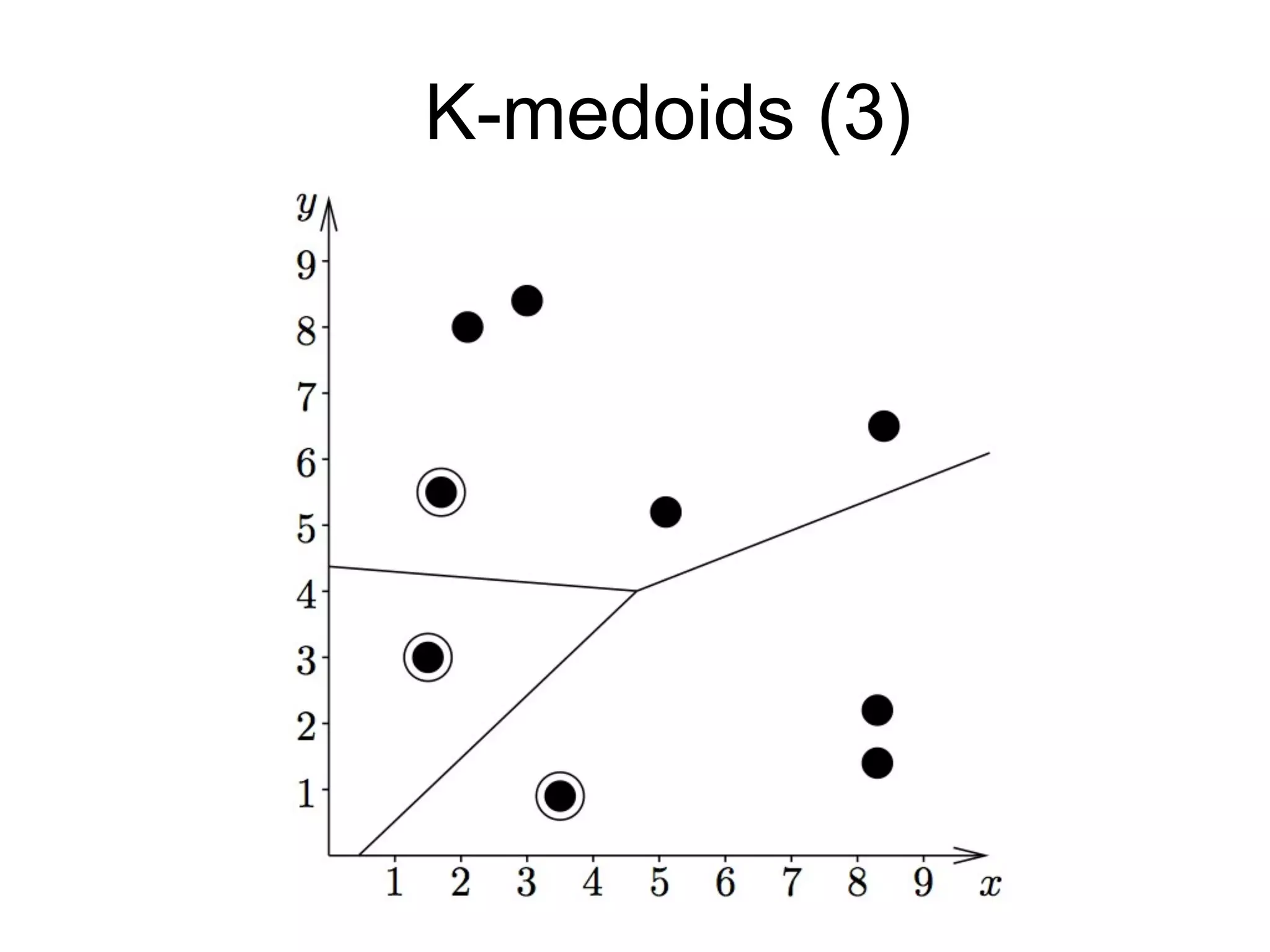
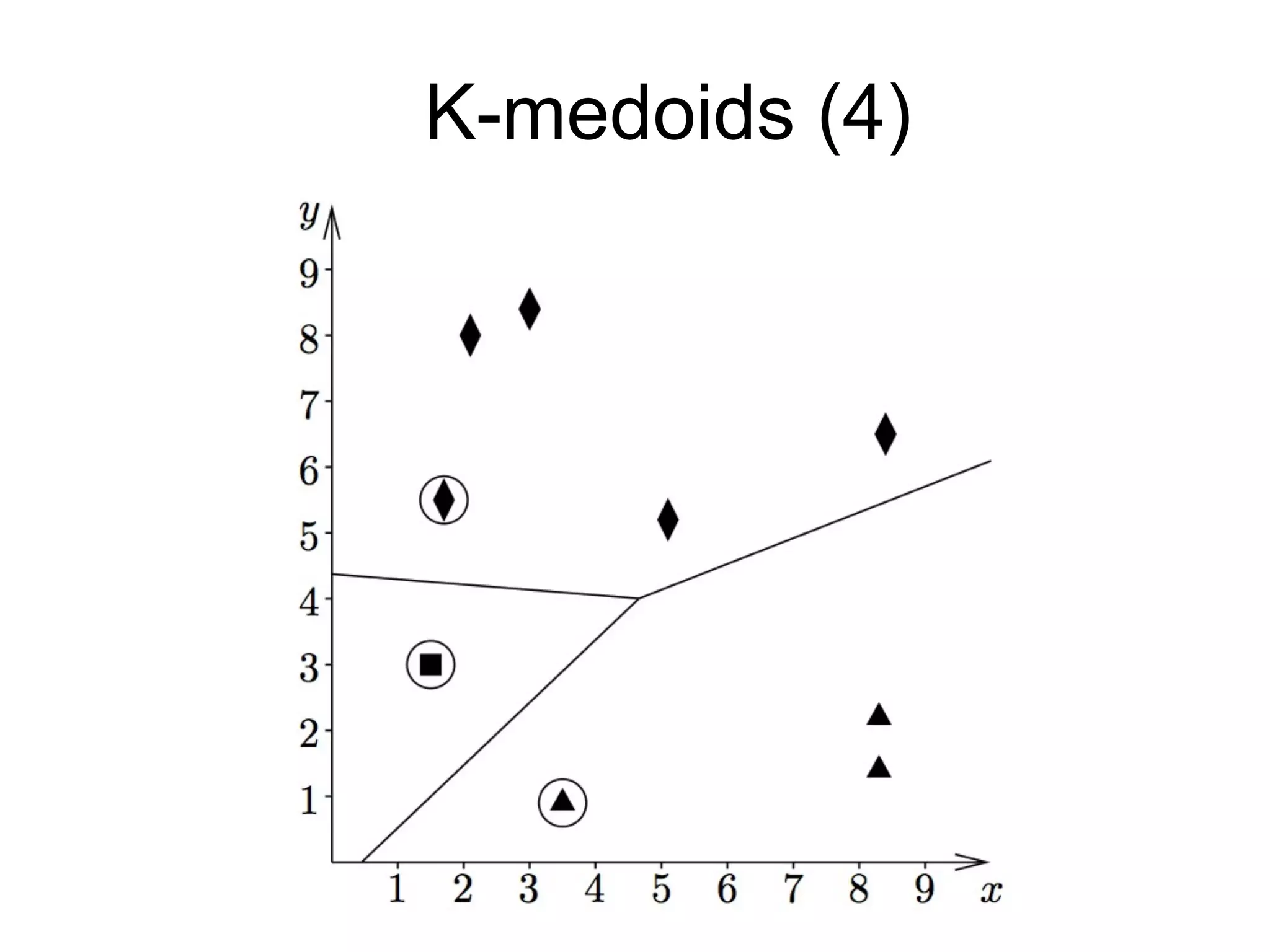
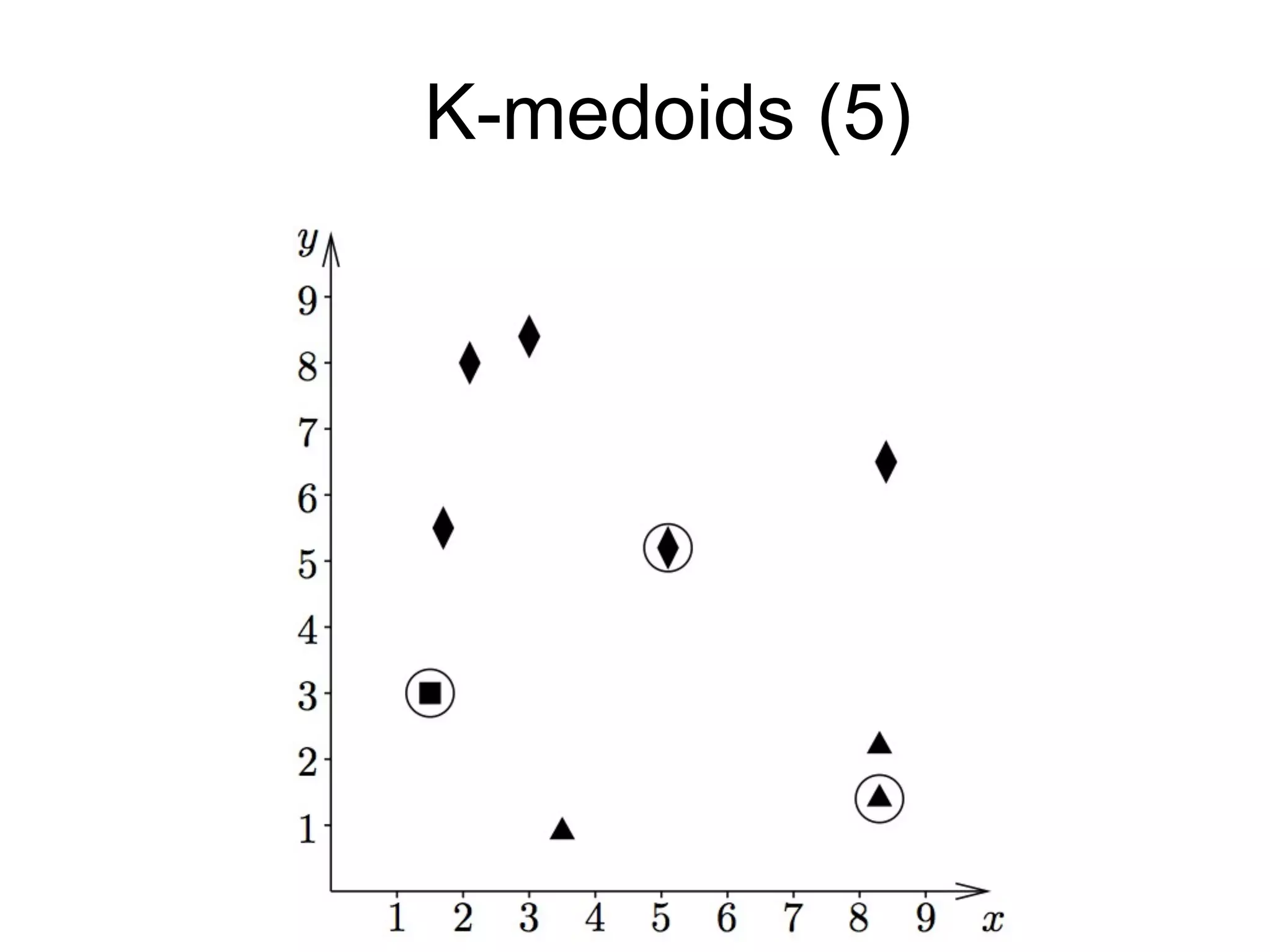
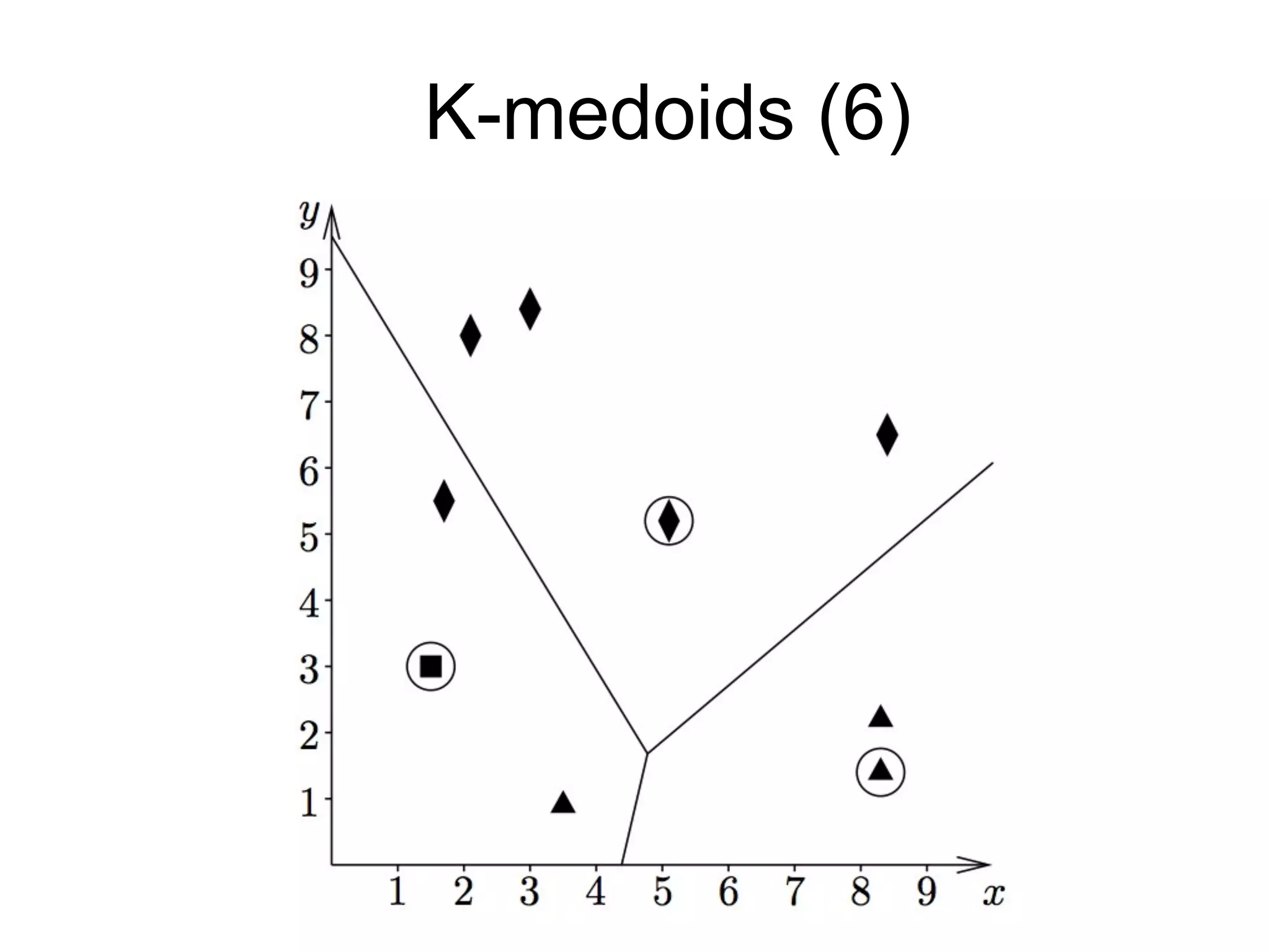
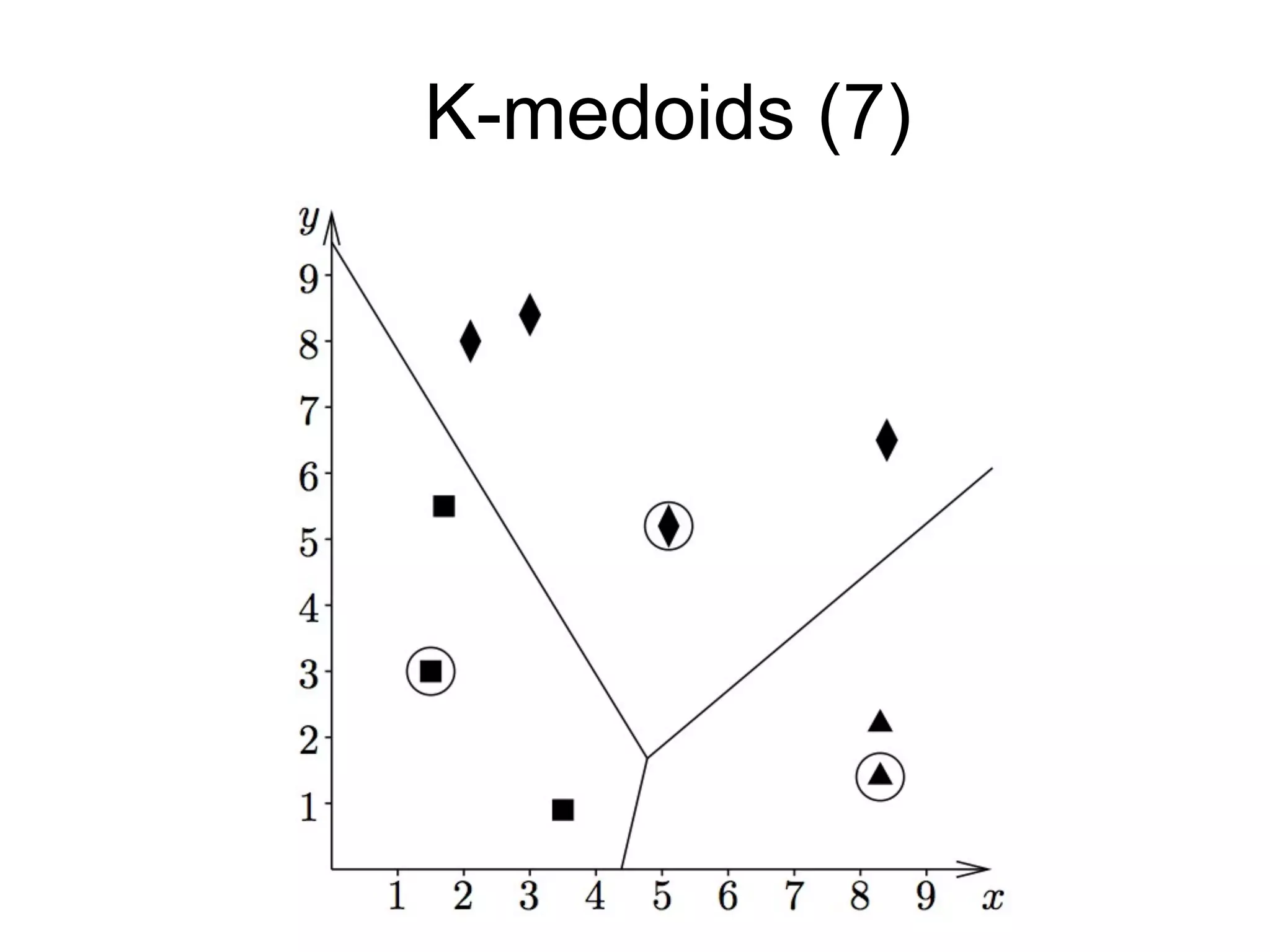
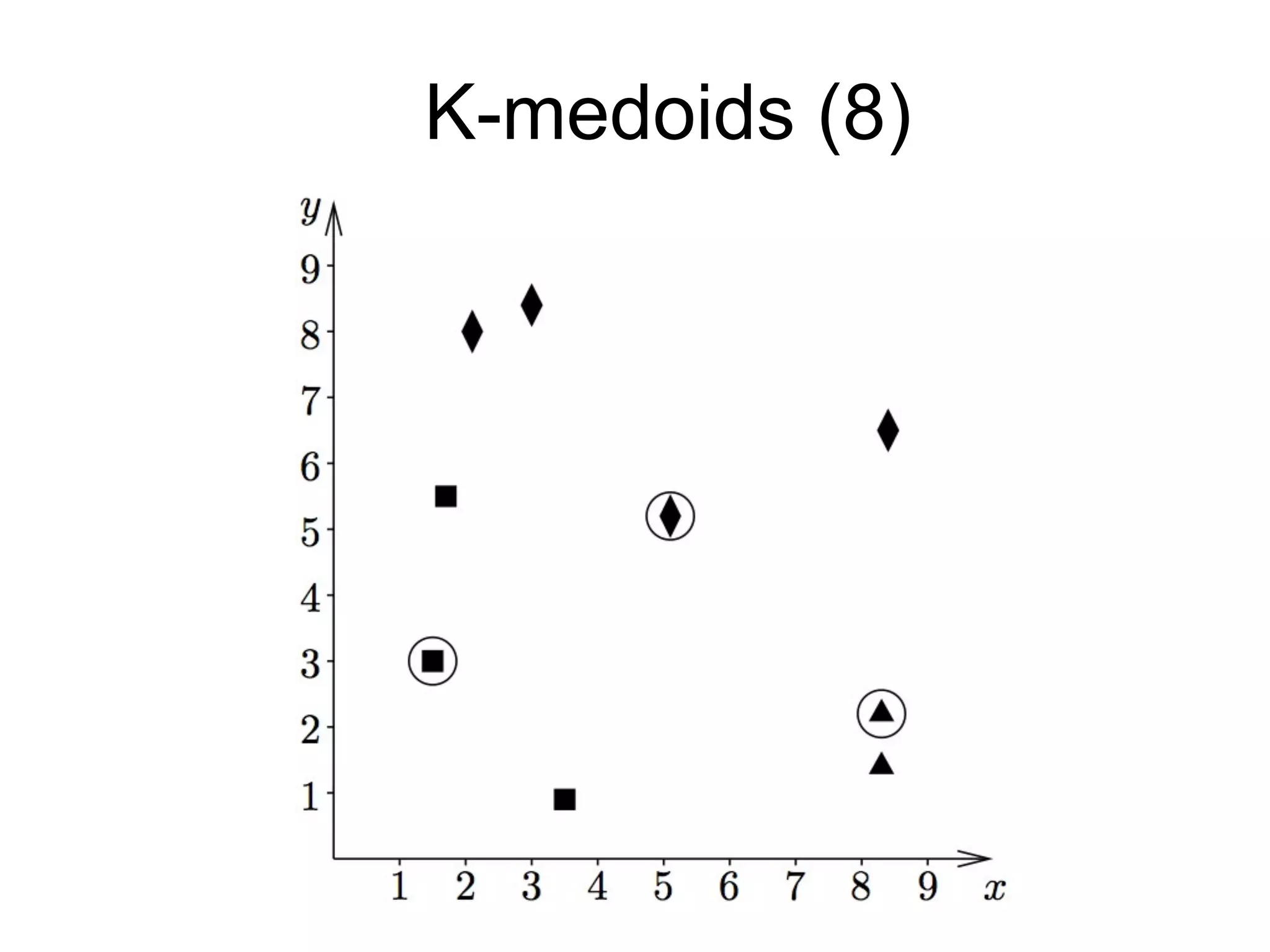
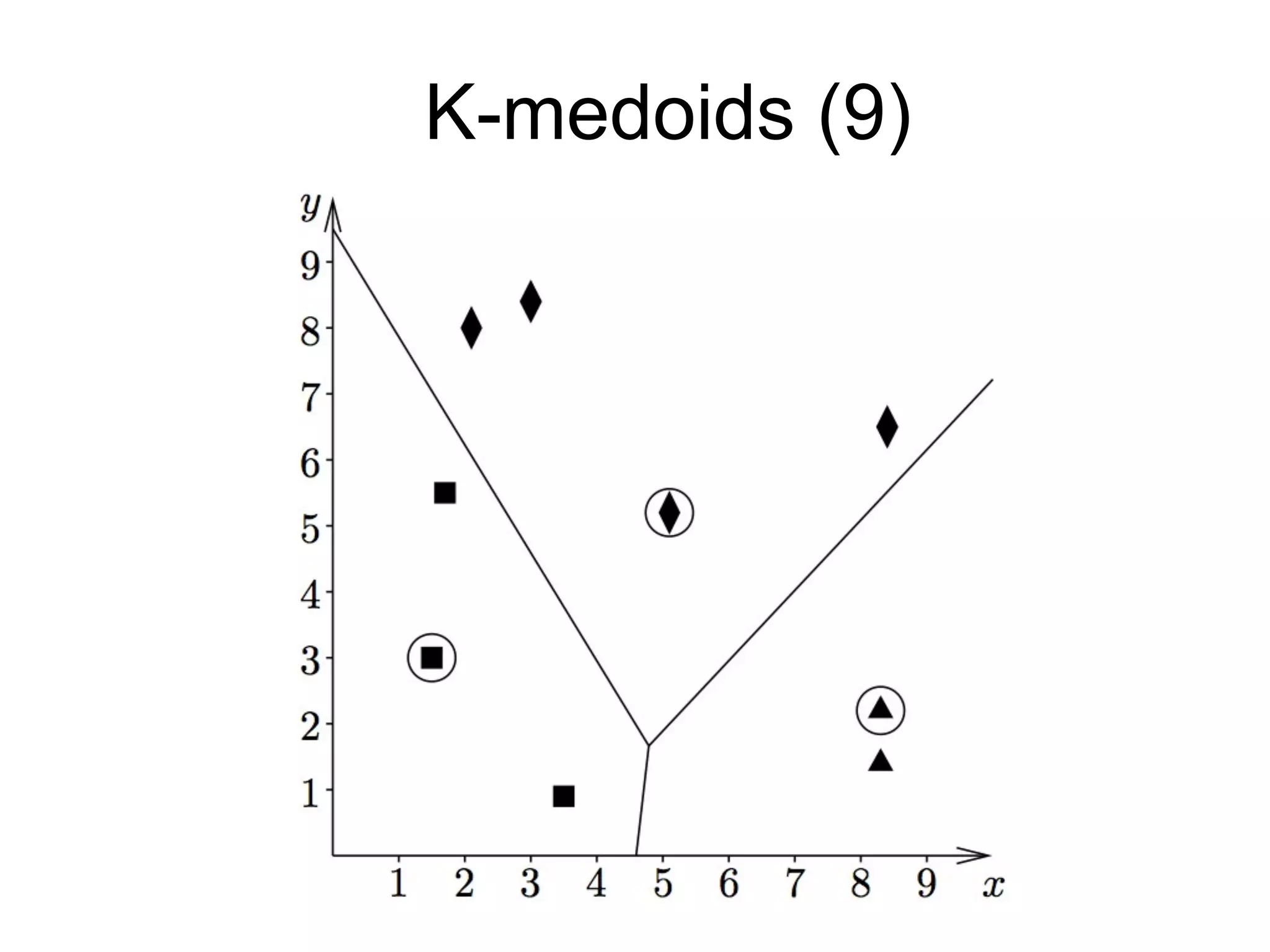
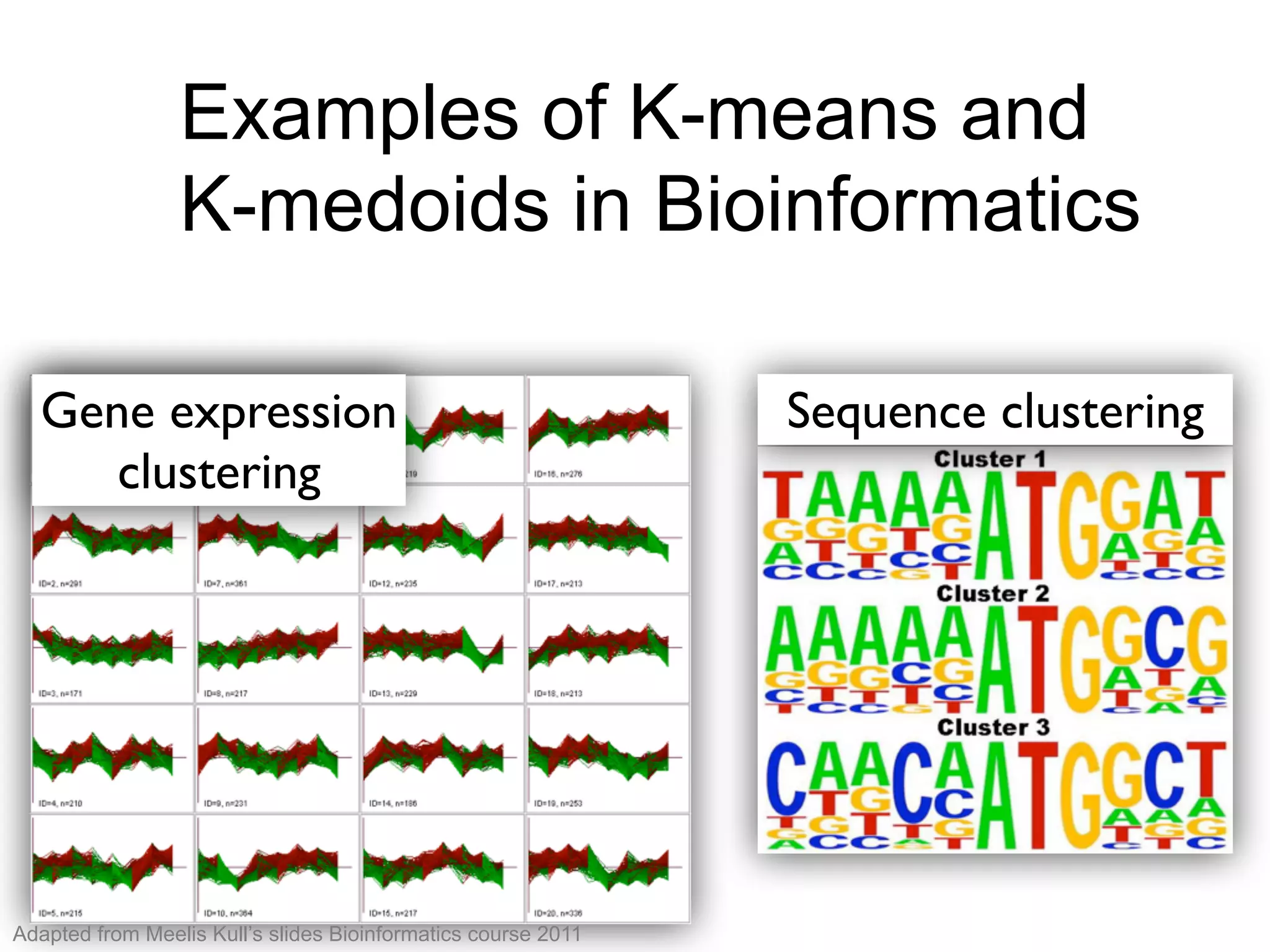
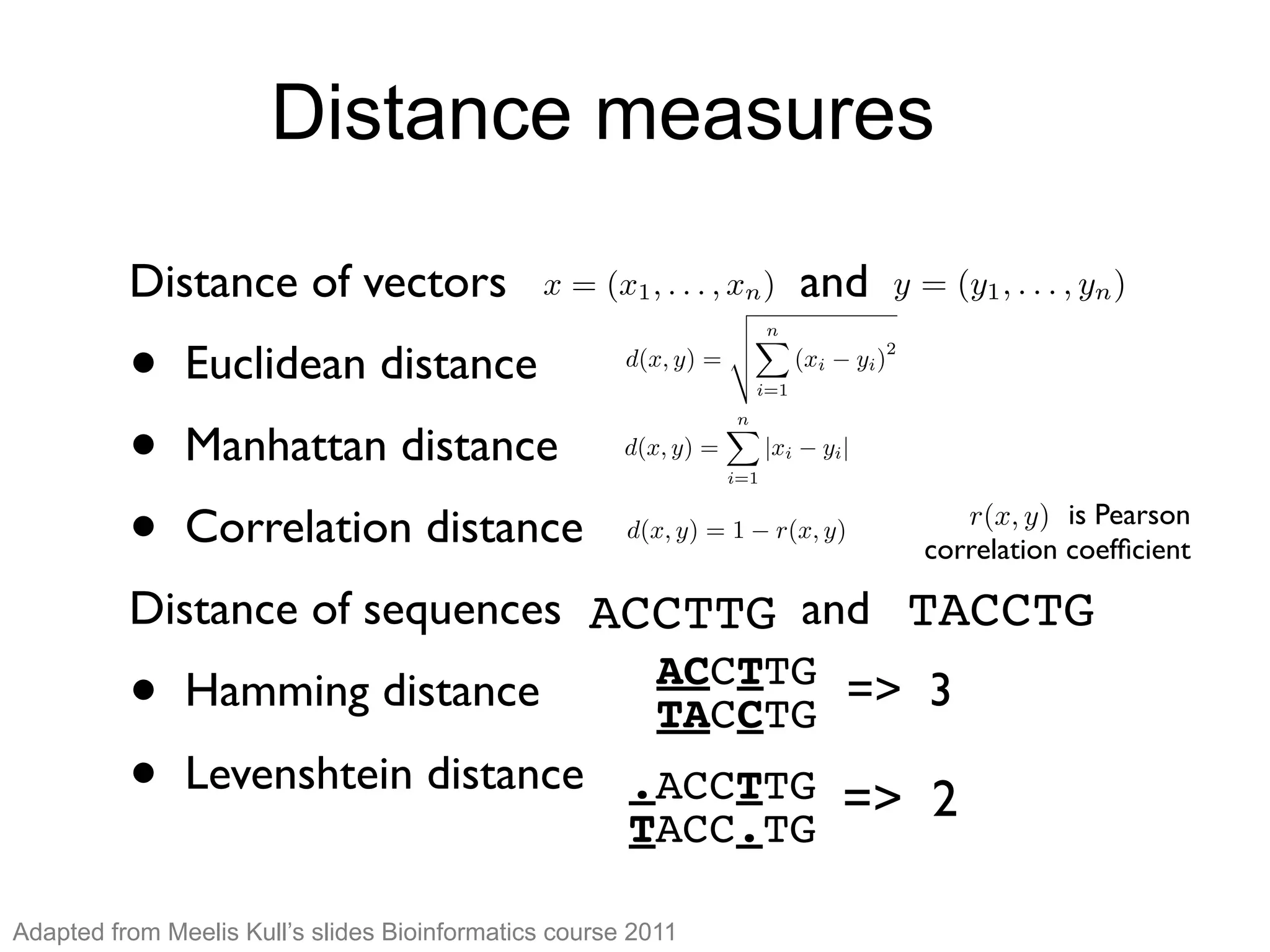
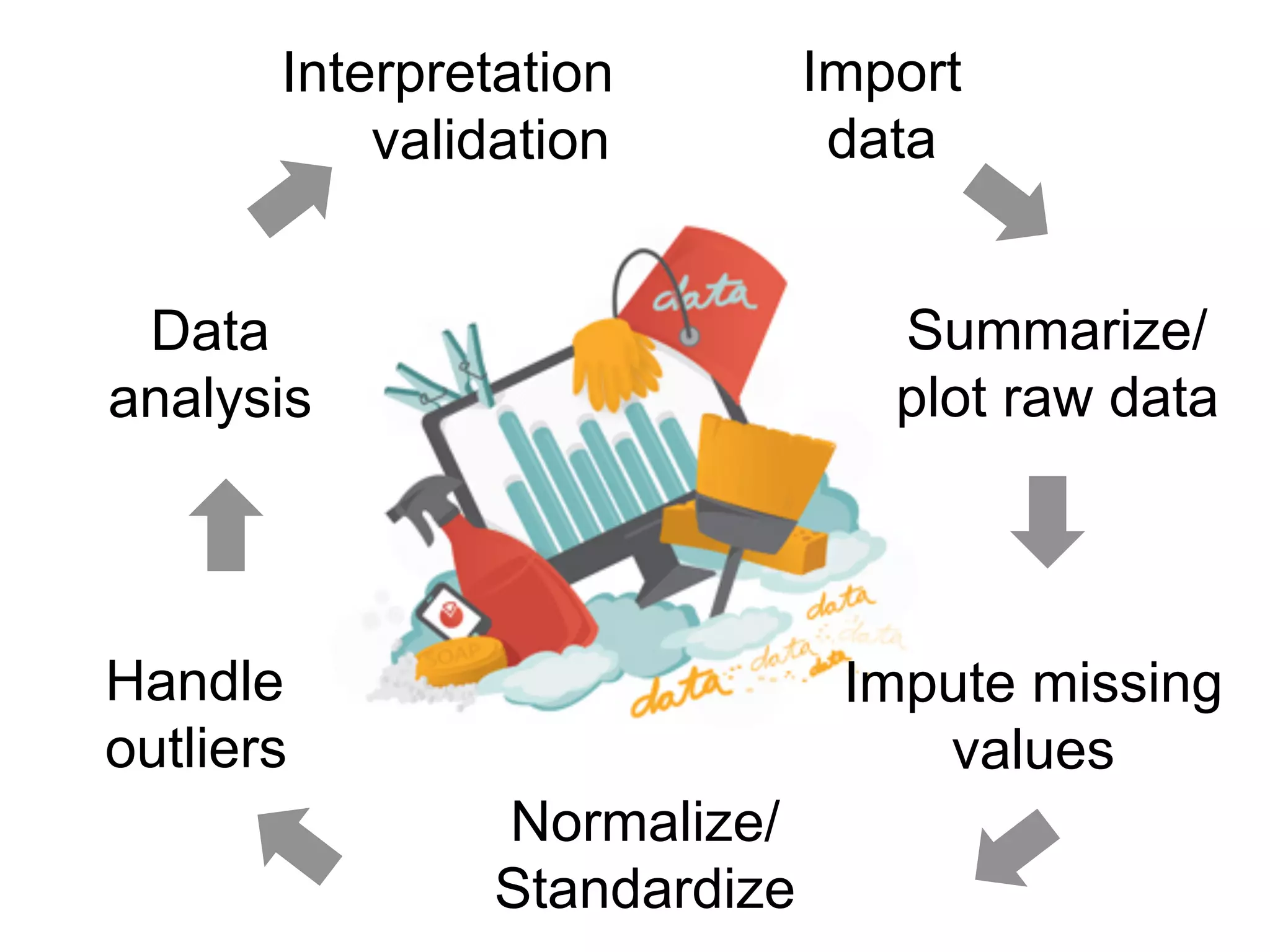
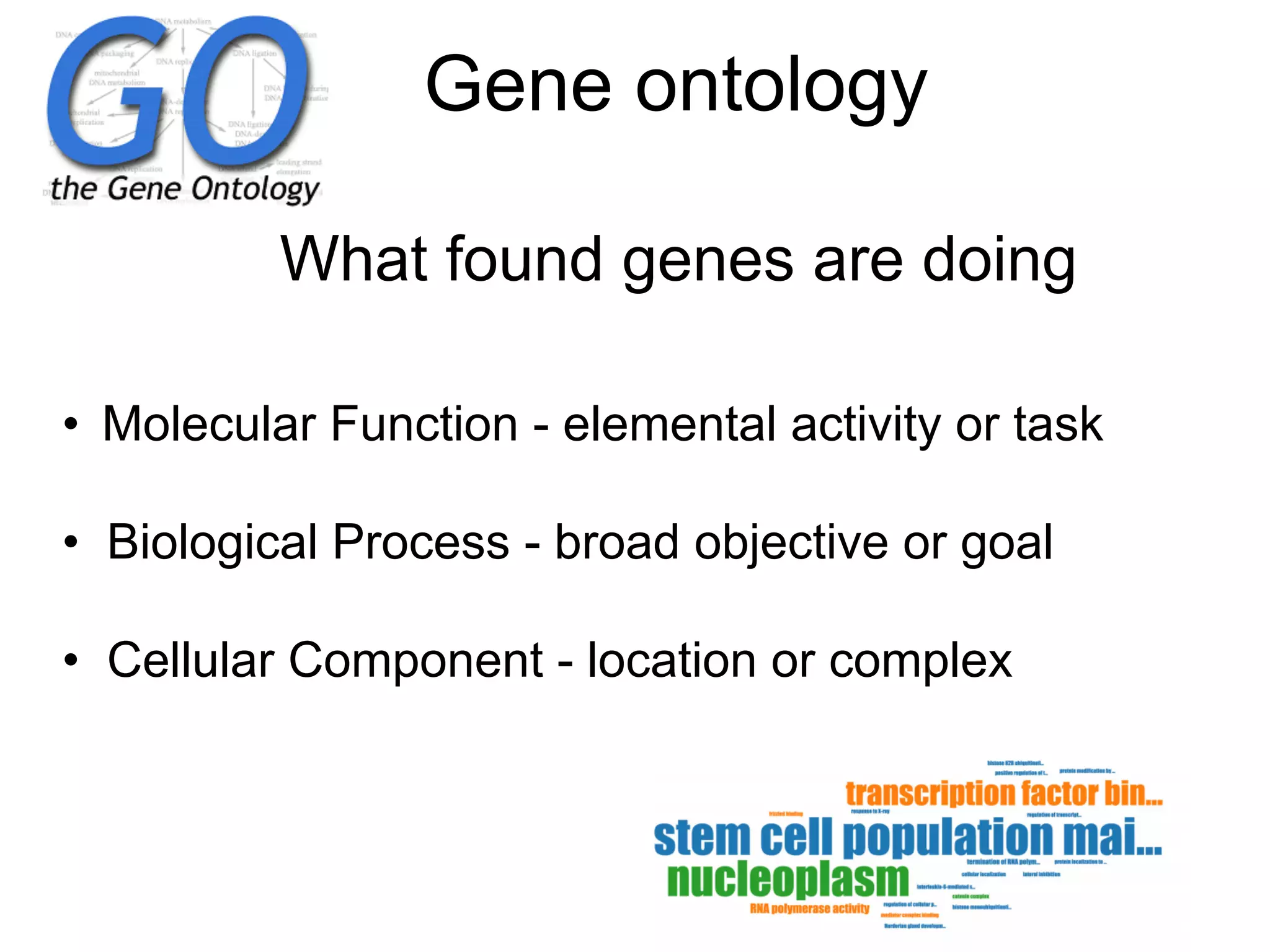
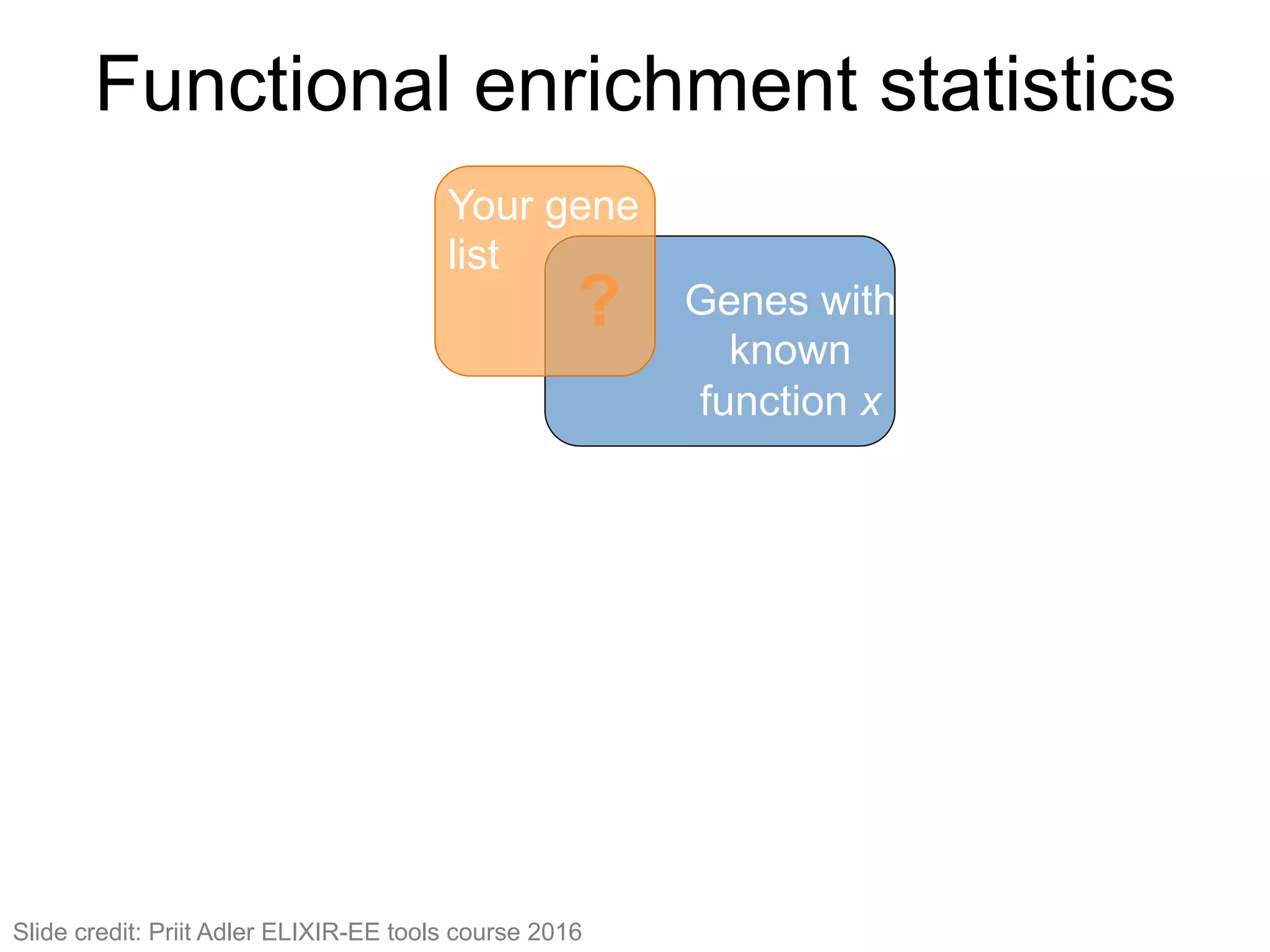
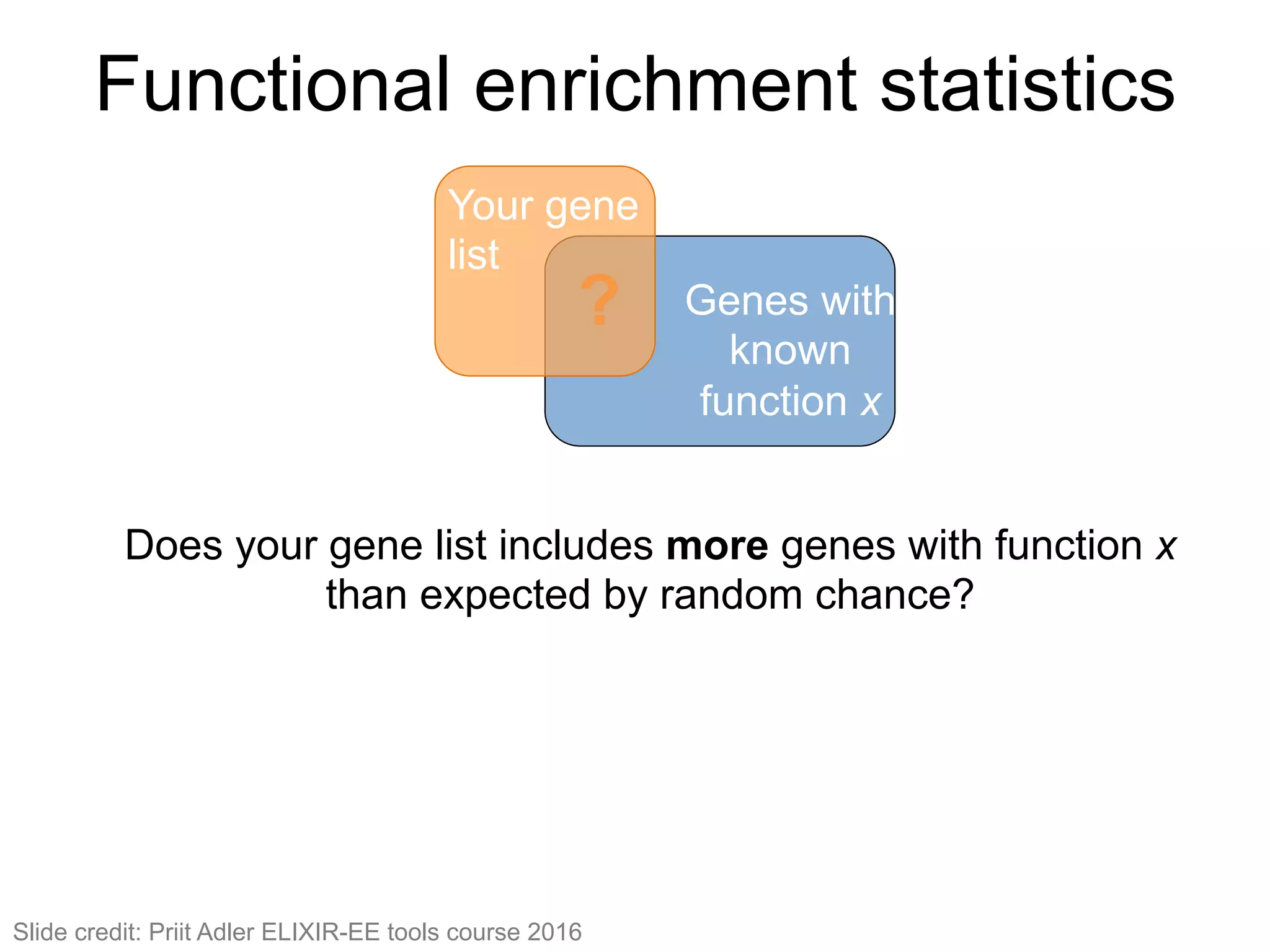
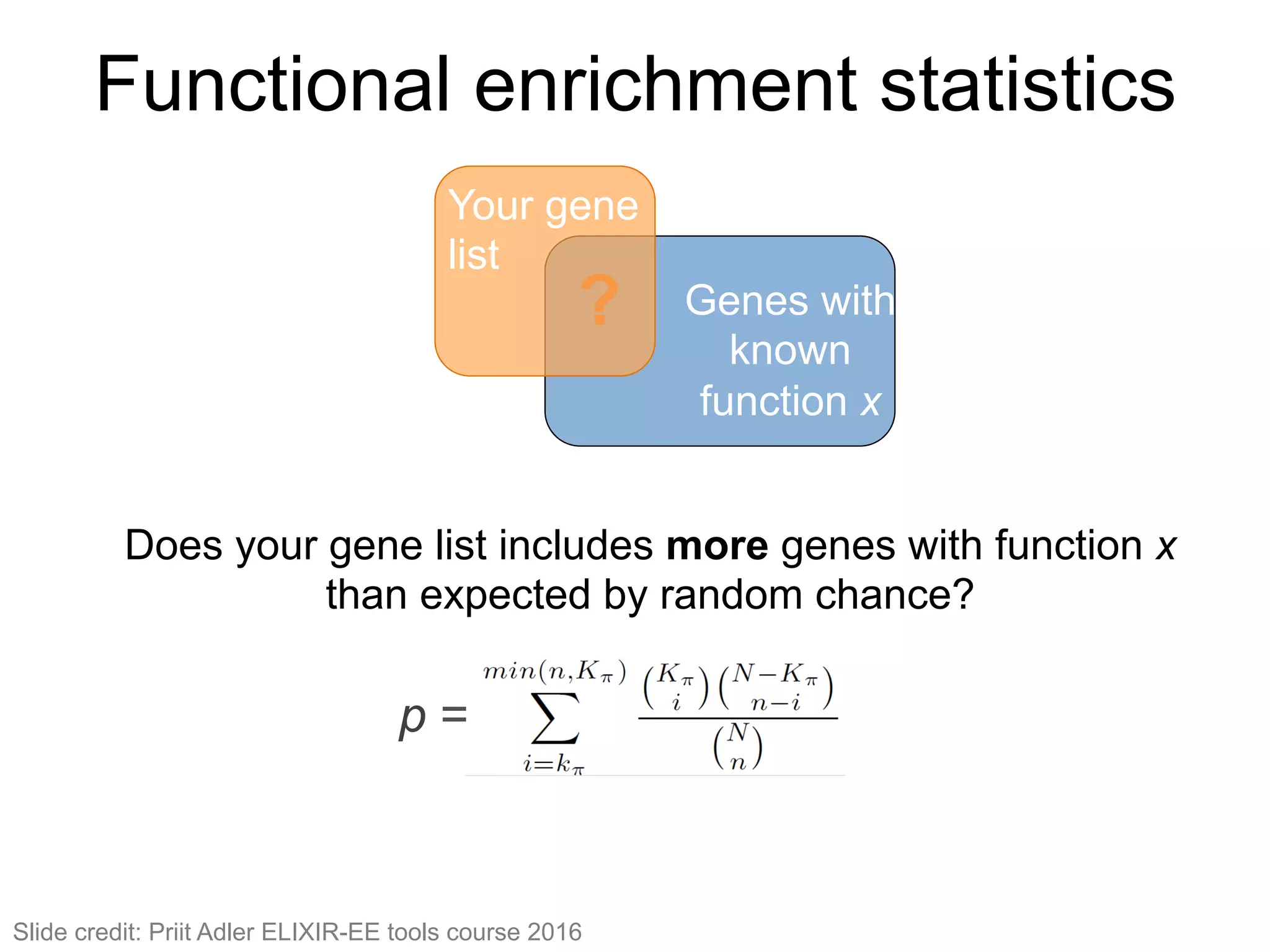
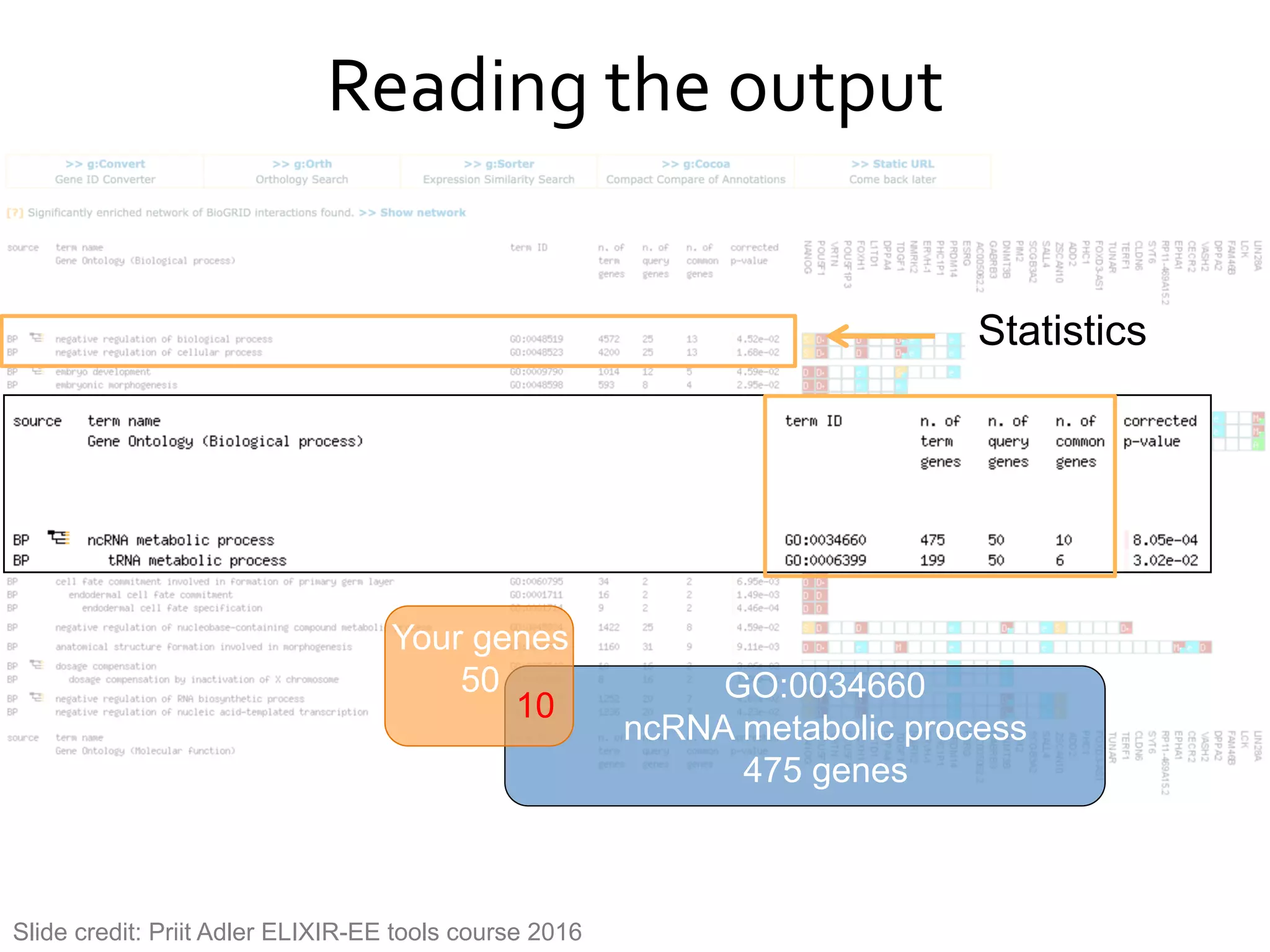
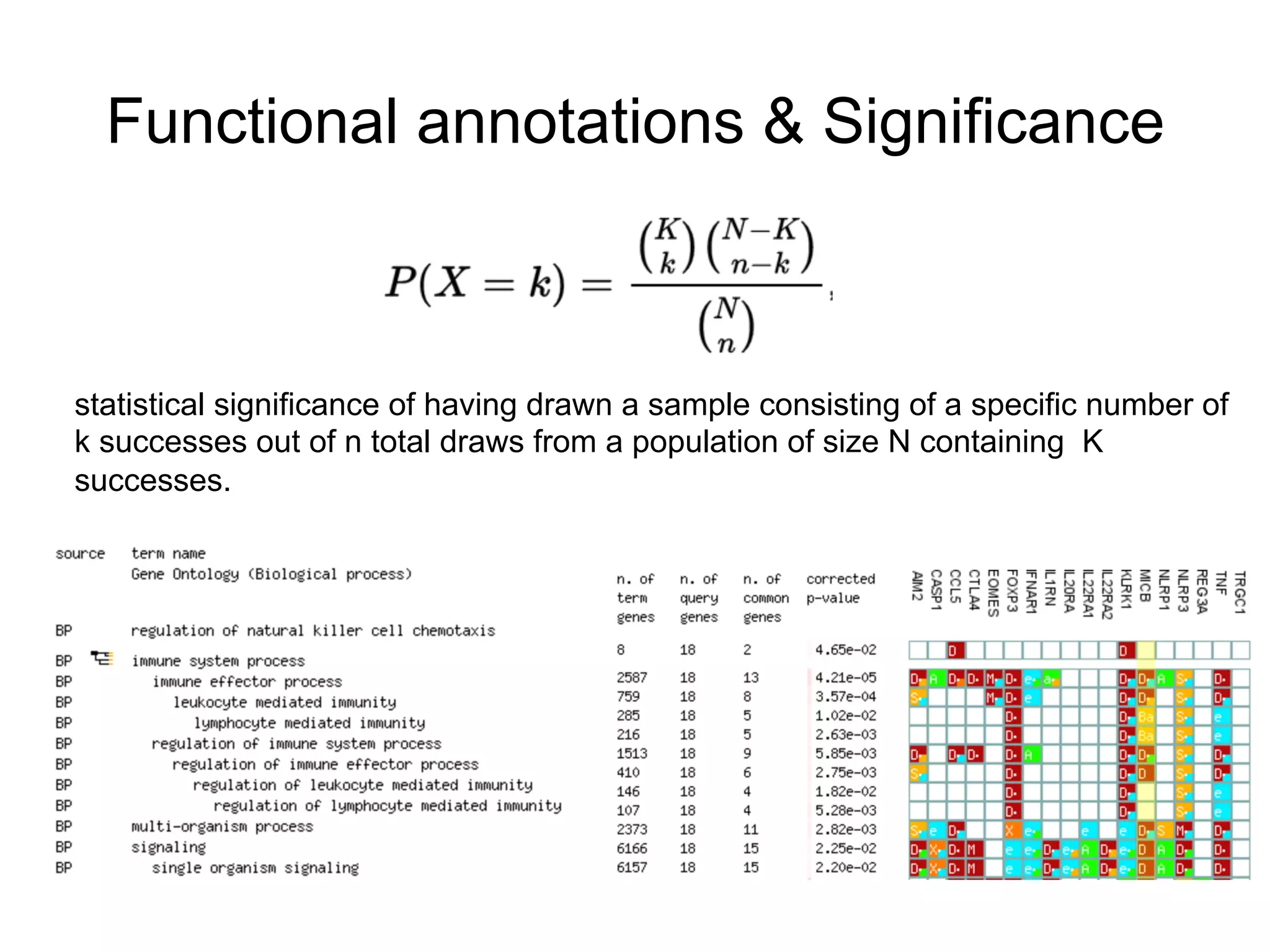
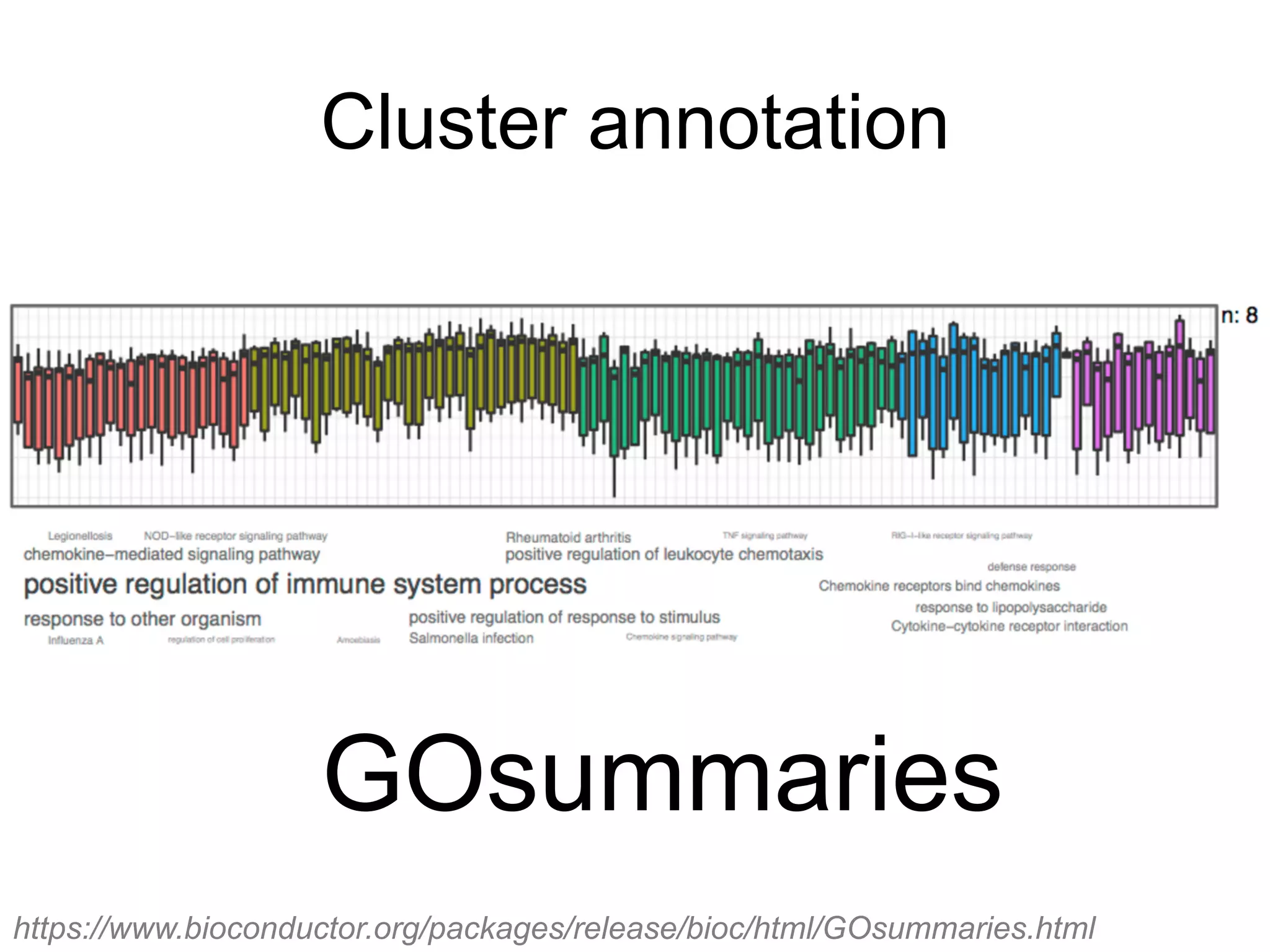
The document provides an overview of data analysis techniques in bioinformatics, focusing on gene expression profiles, data preprocessing, and methods for handling missing values and normalization. It discusses clustering methods, such as hierarchical clustering and k-means, along with techniques for visualizing and interpreting biological data. Additionally, the document emphasizes the importance of addressing technical biases and batch effects in experimental design.