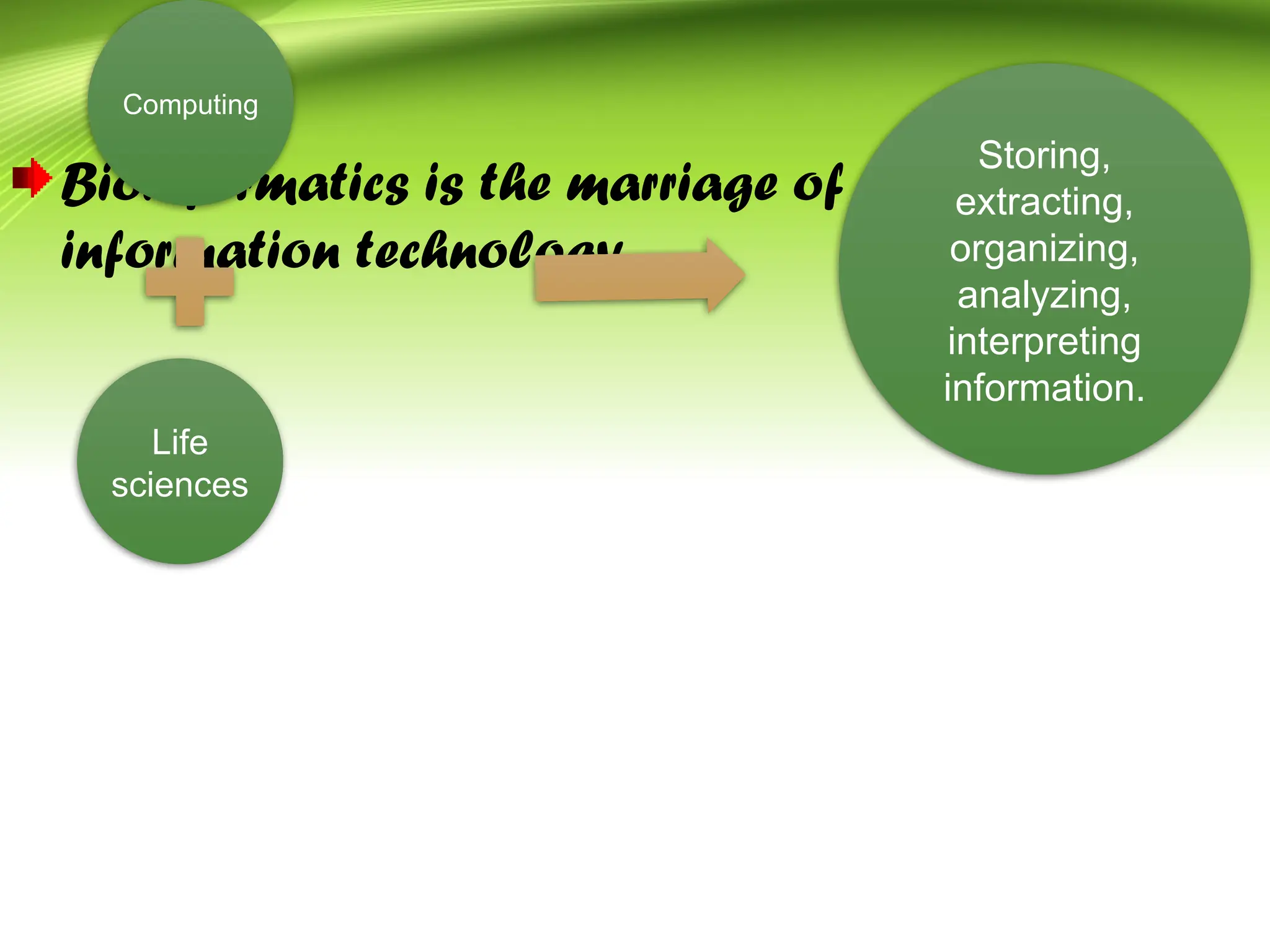
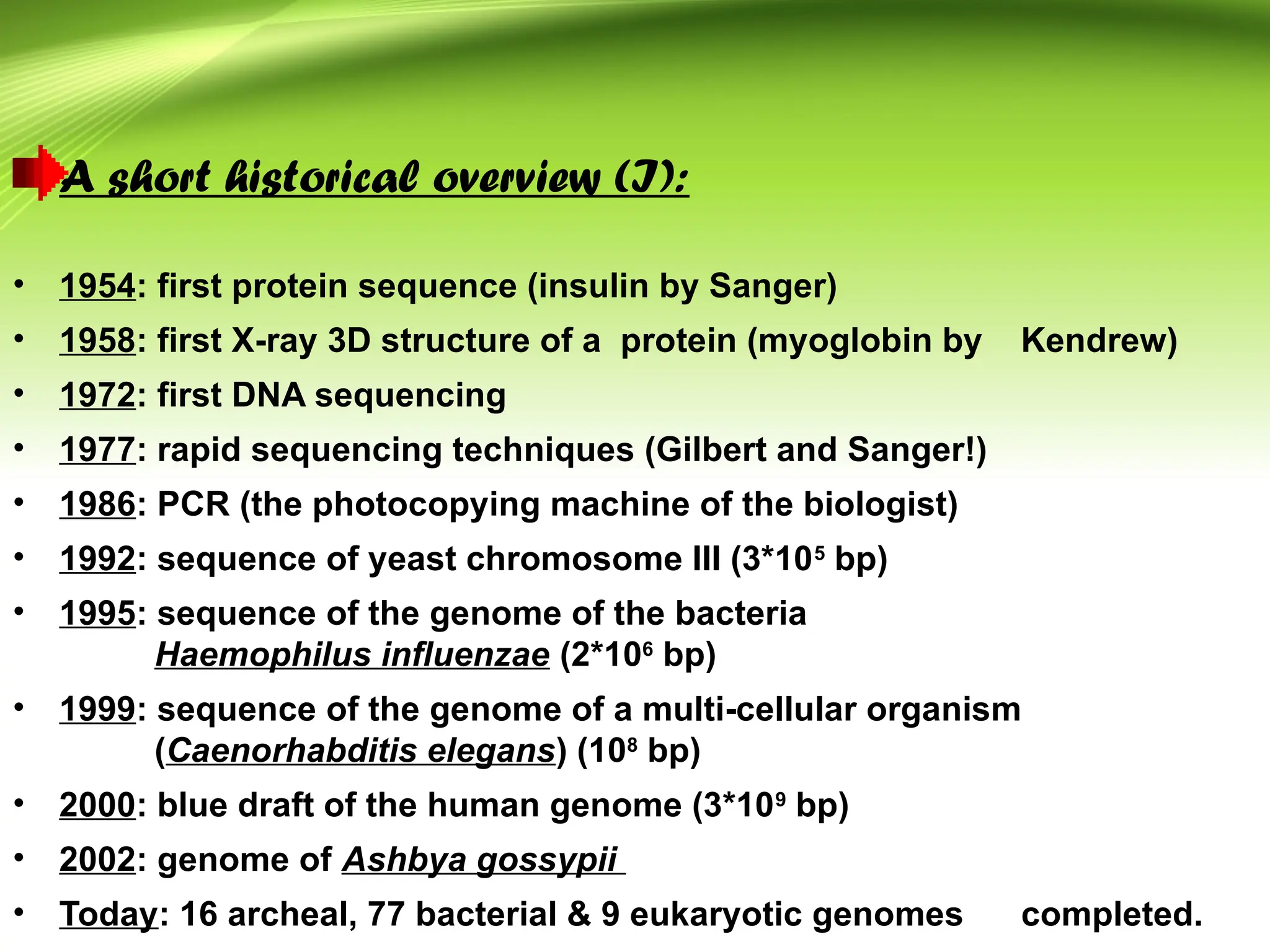
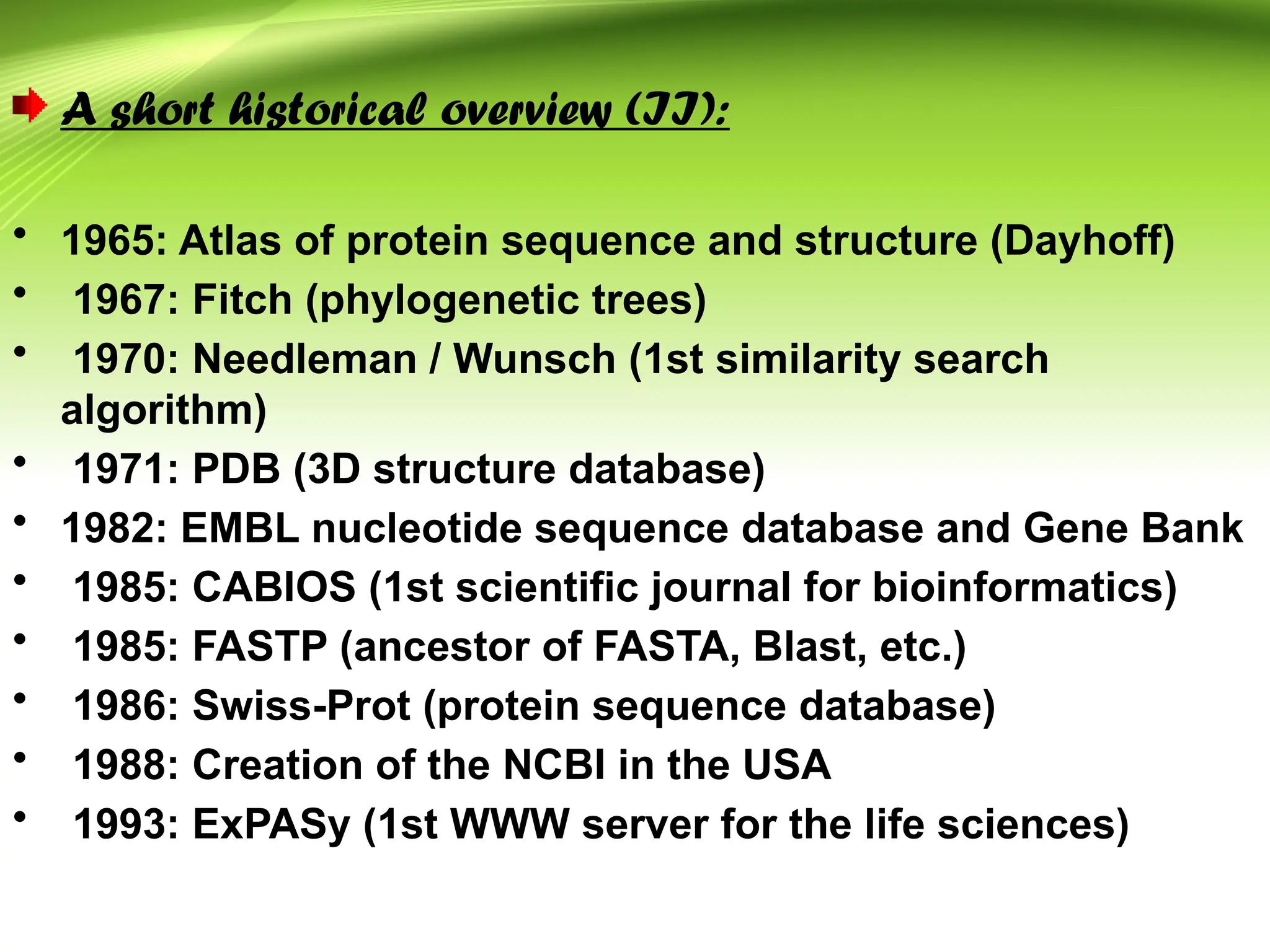
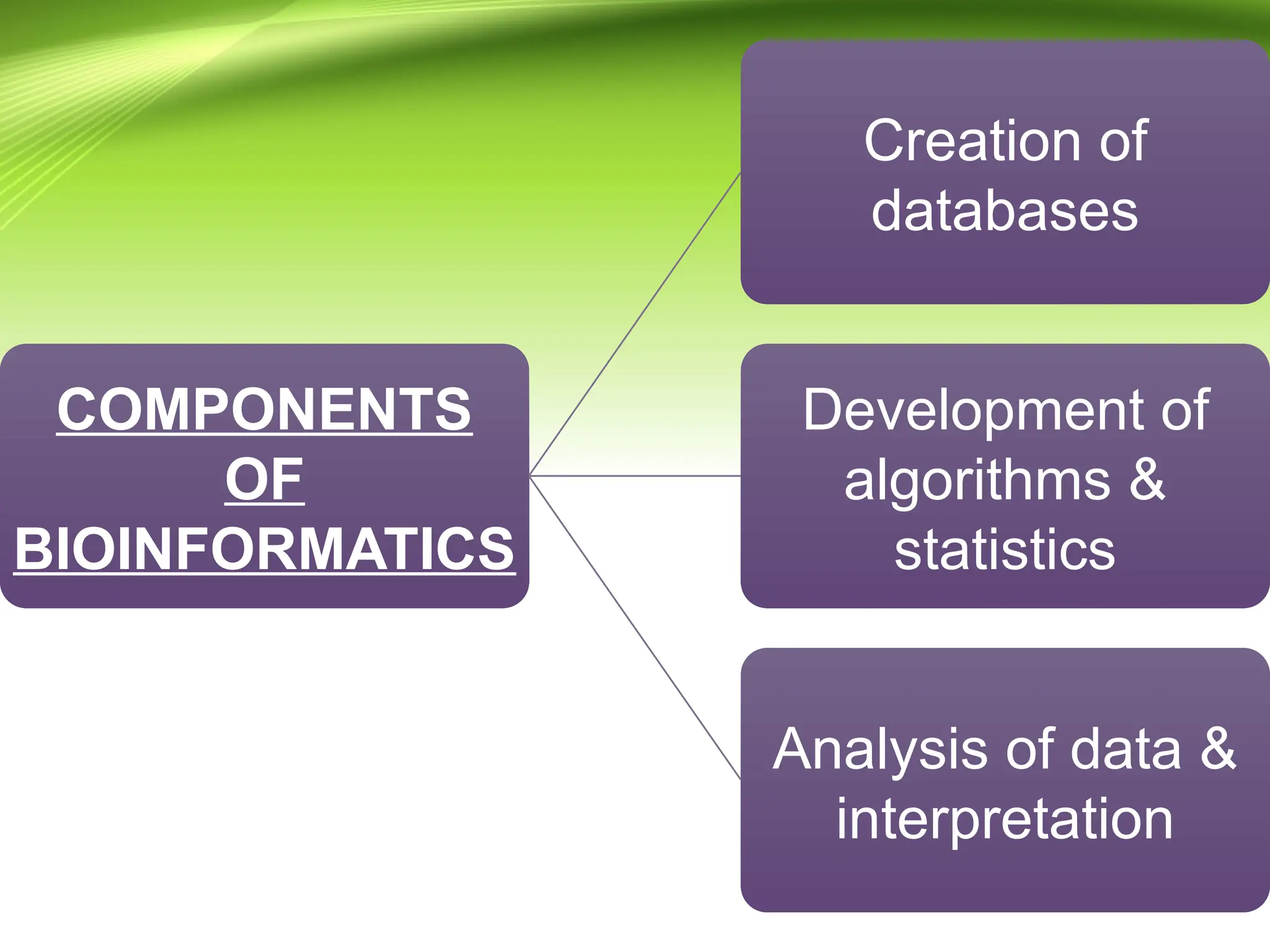
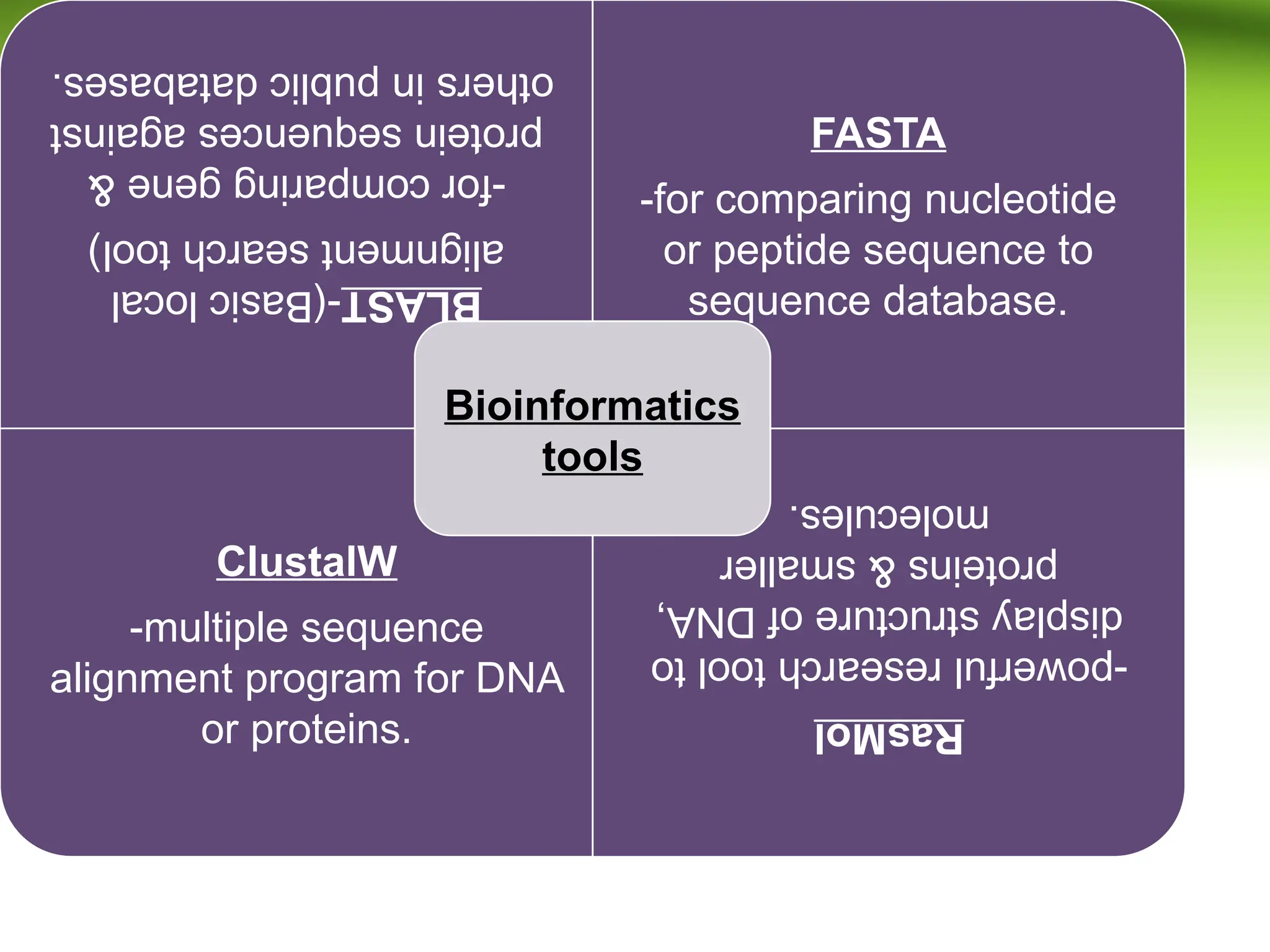
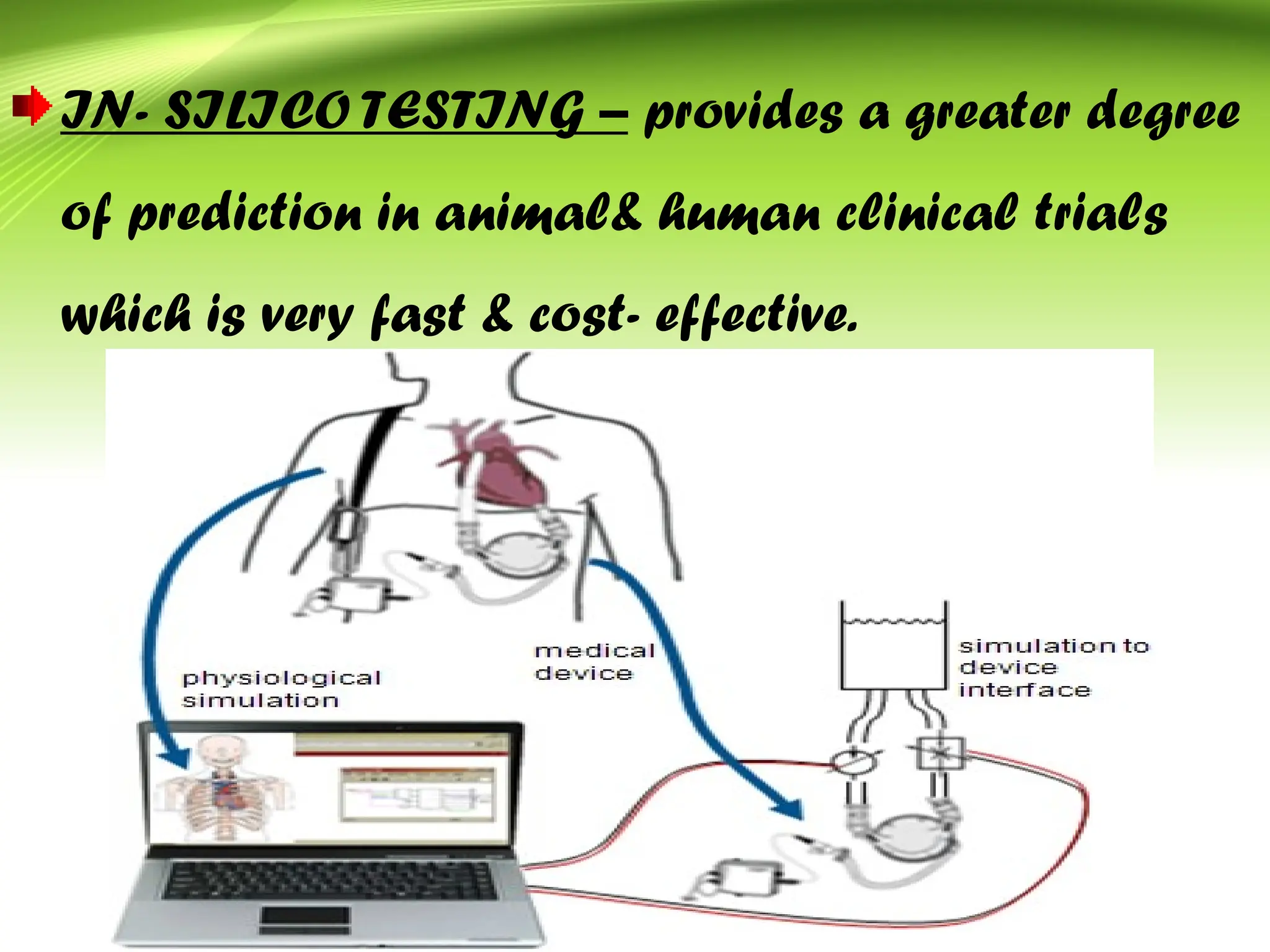
Bioinformatics integrates biology and information technology to analyze and interpret biological data, with applications in various fields such as molecular medicine, genomics, and drug design. Its historical development spans key milestones in DNA sequencing and database creation, reflecting its growing importance in scientific research and pharmaceutical industries. The field continues to evolve, providing tools and techniques that enhance our understanding of biological systems and contribute to advancements in personalized medicine and agricultural improvements.