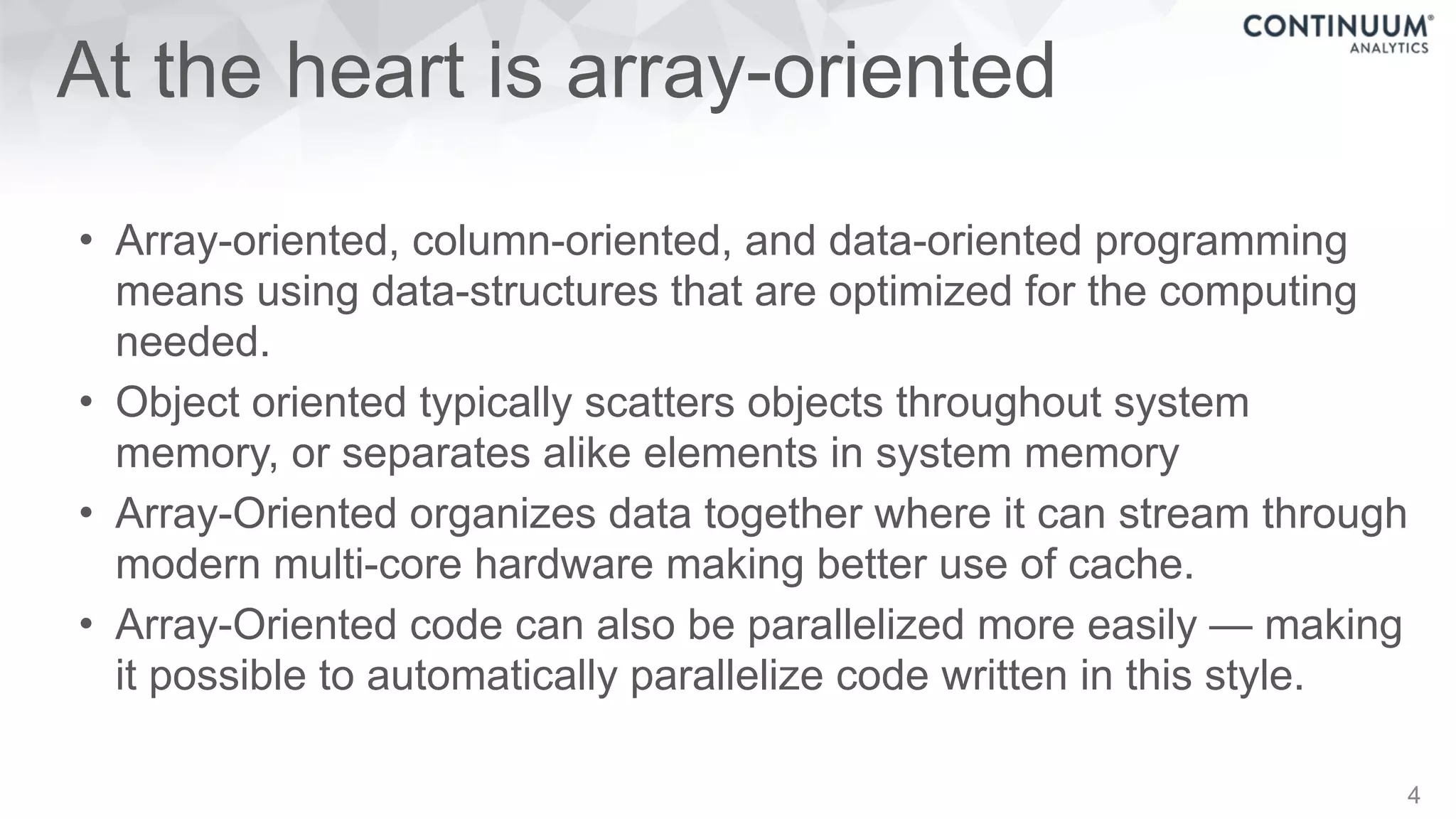
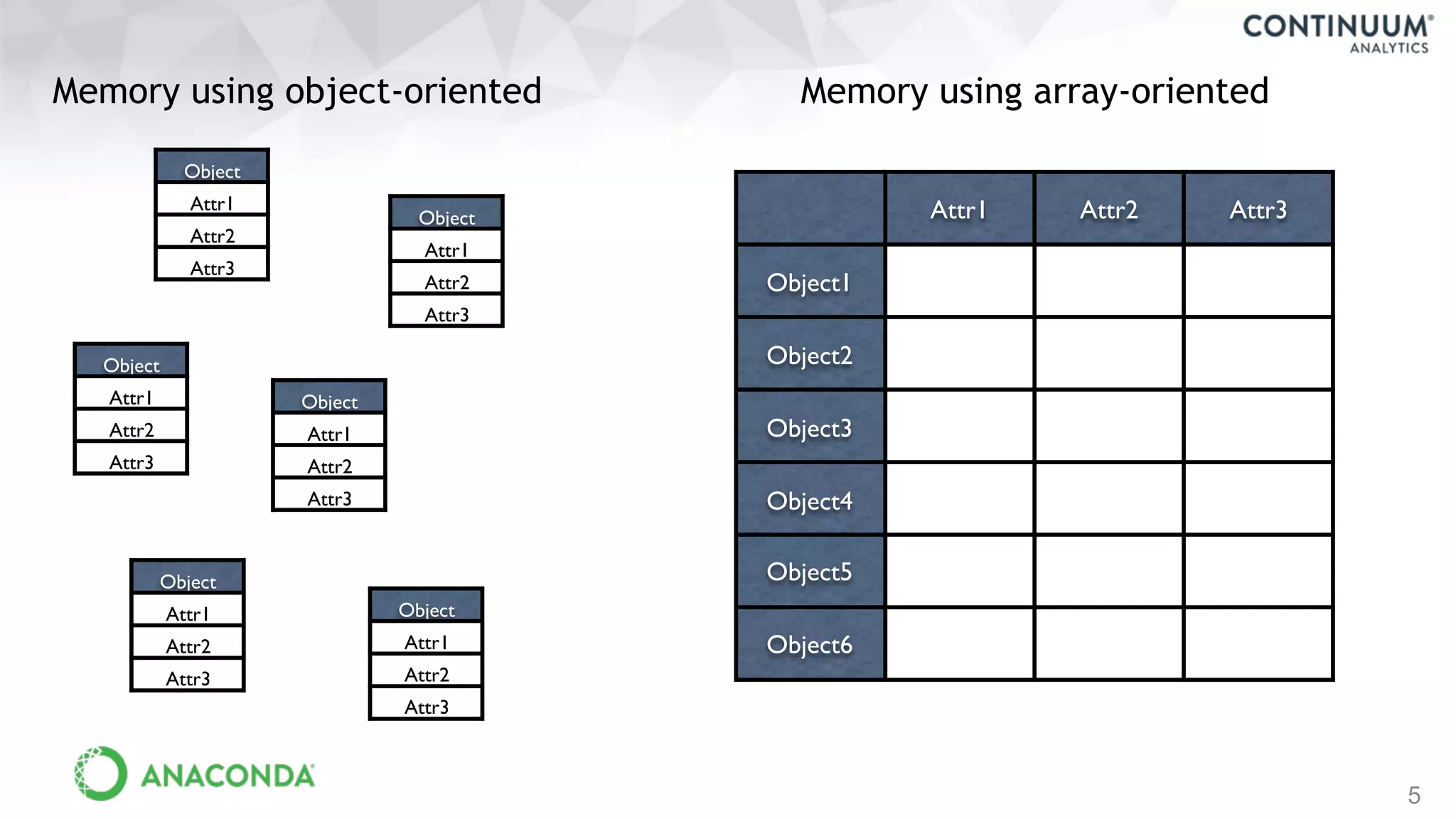
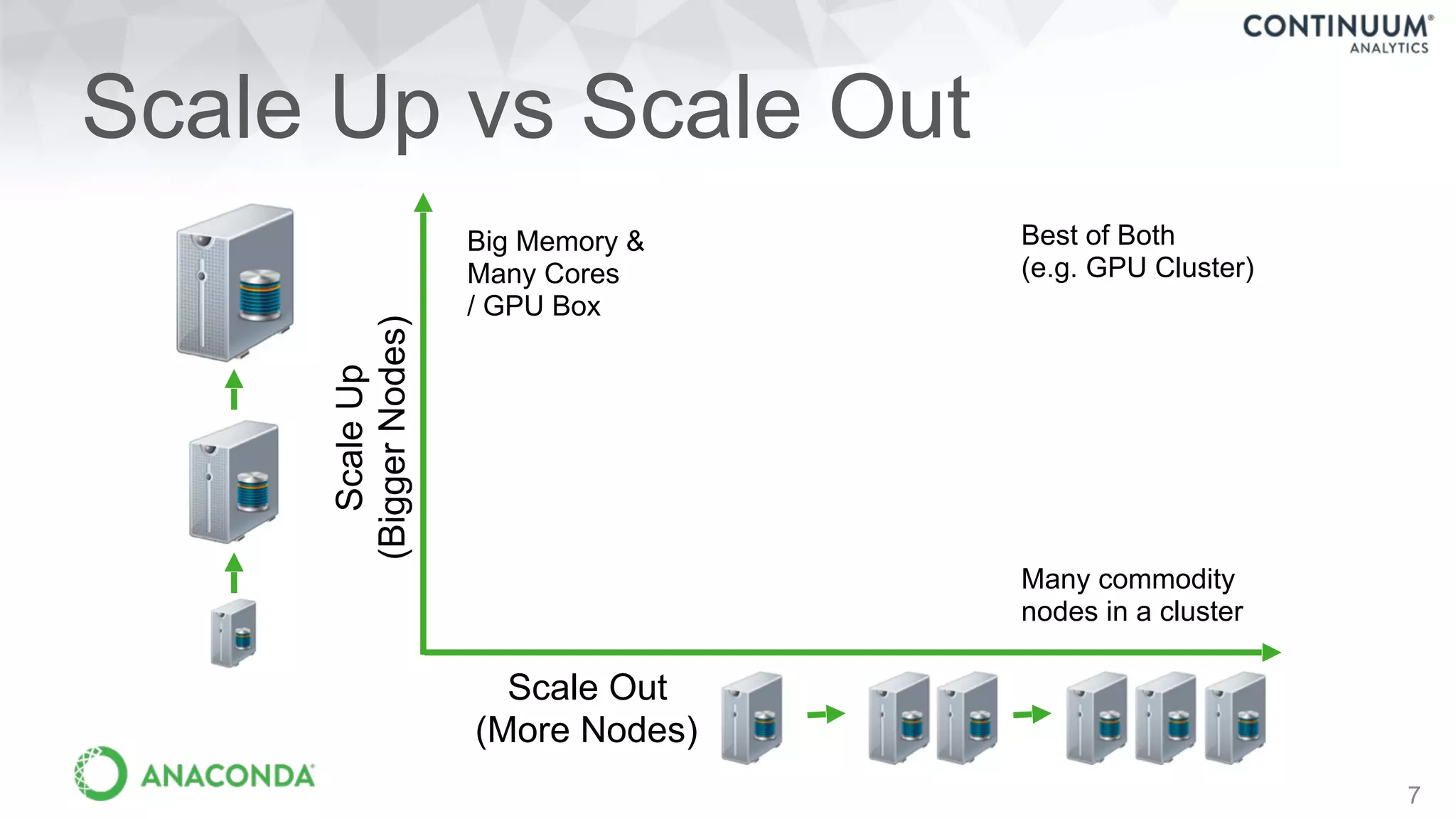
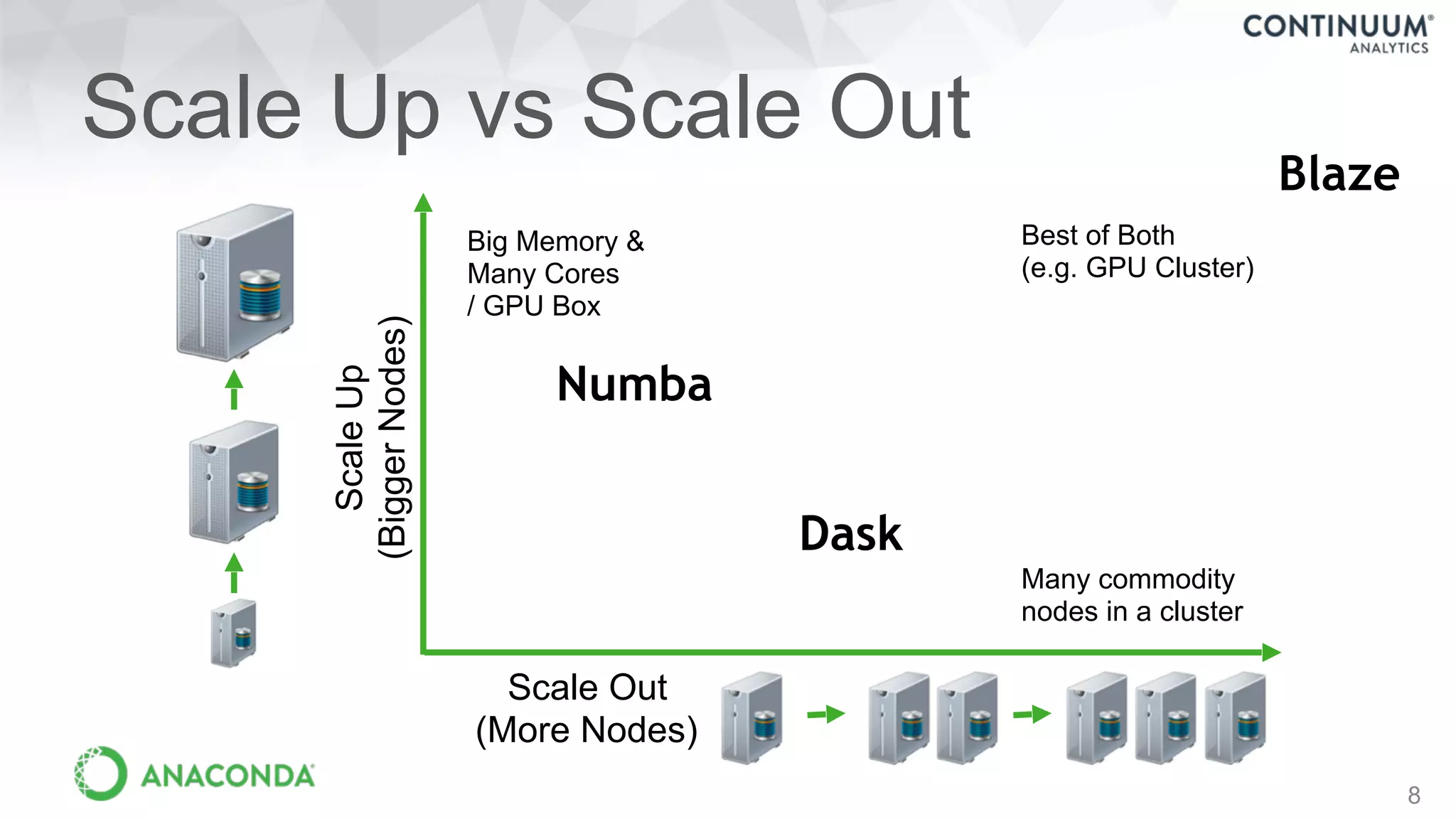
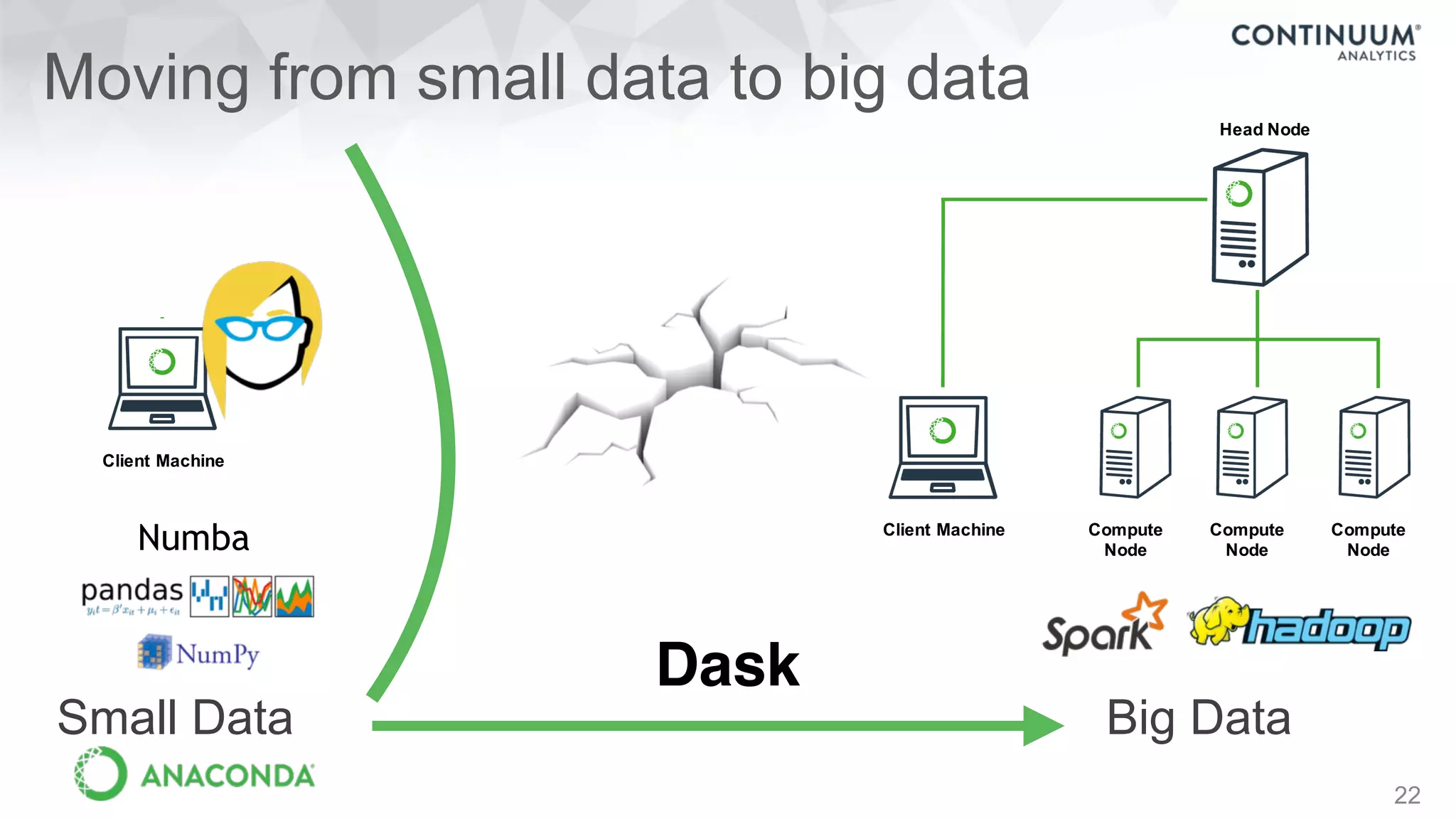
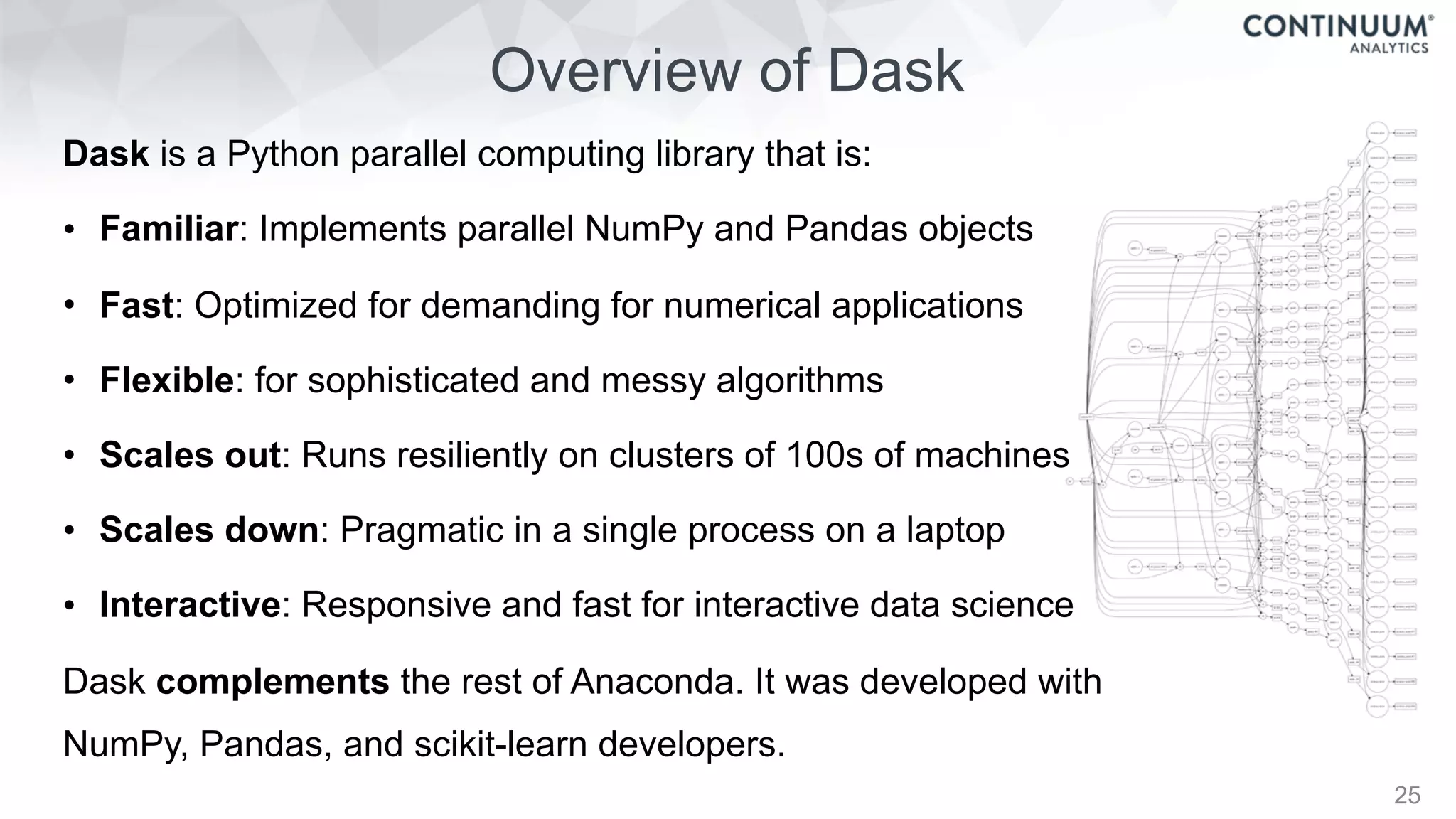
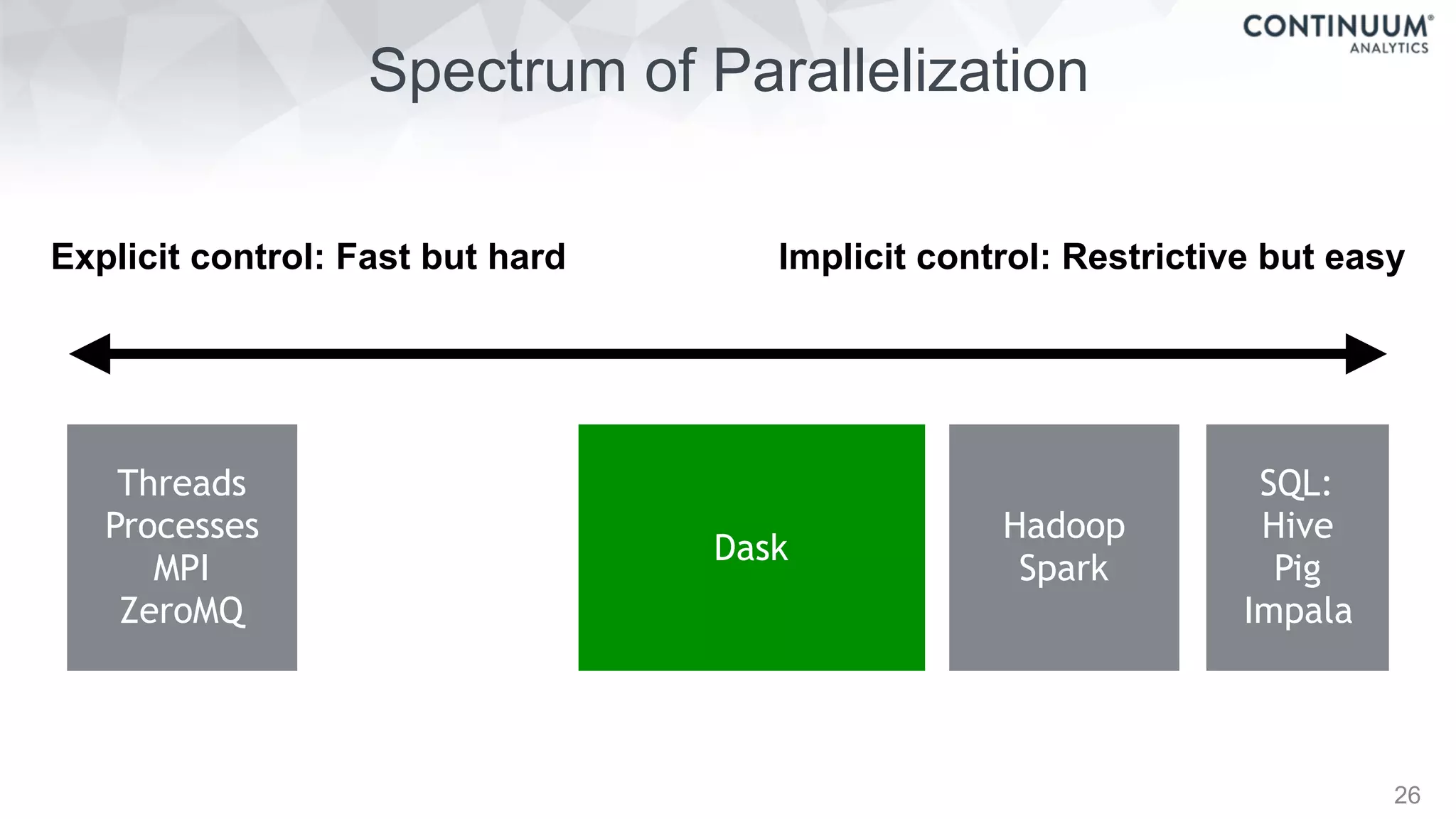
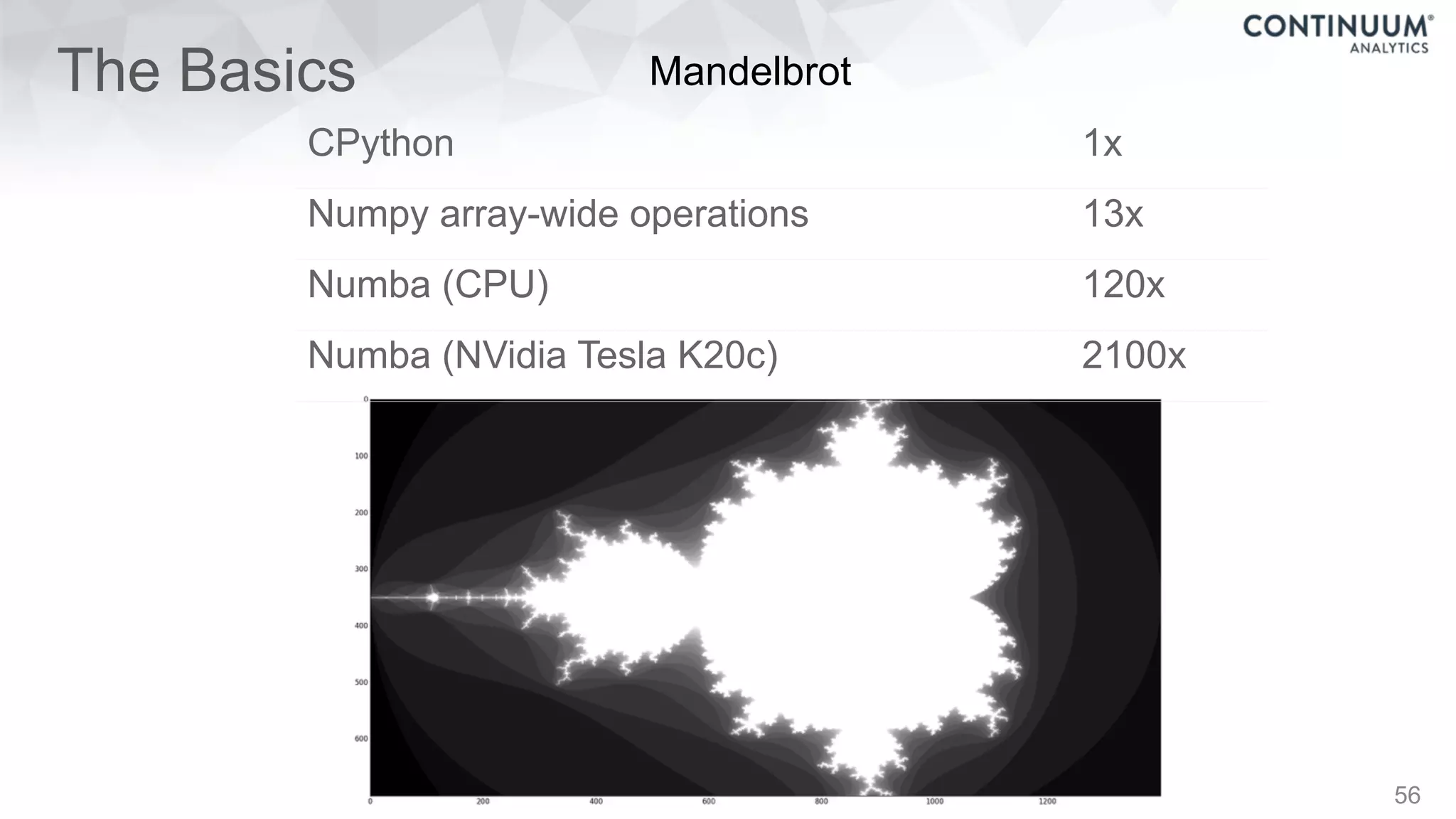
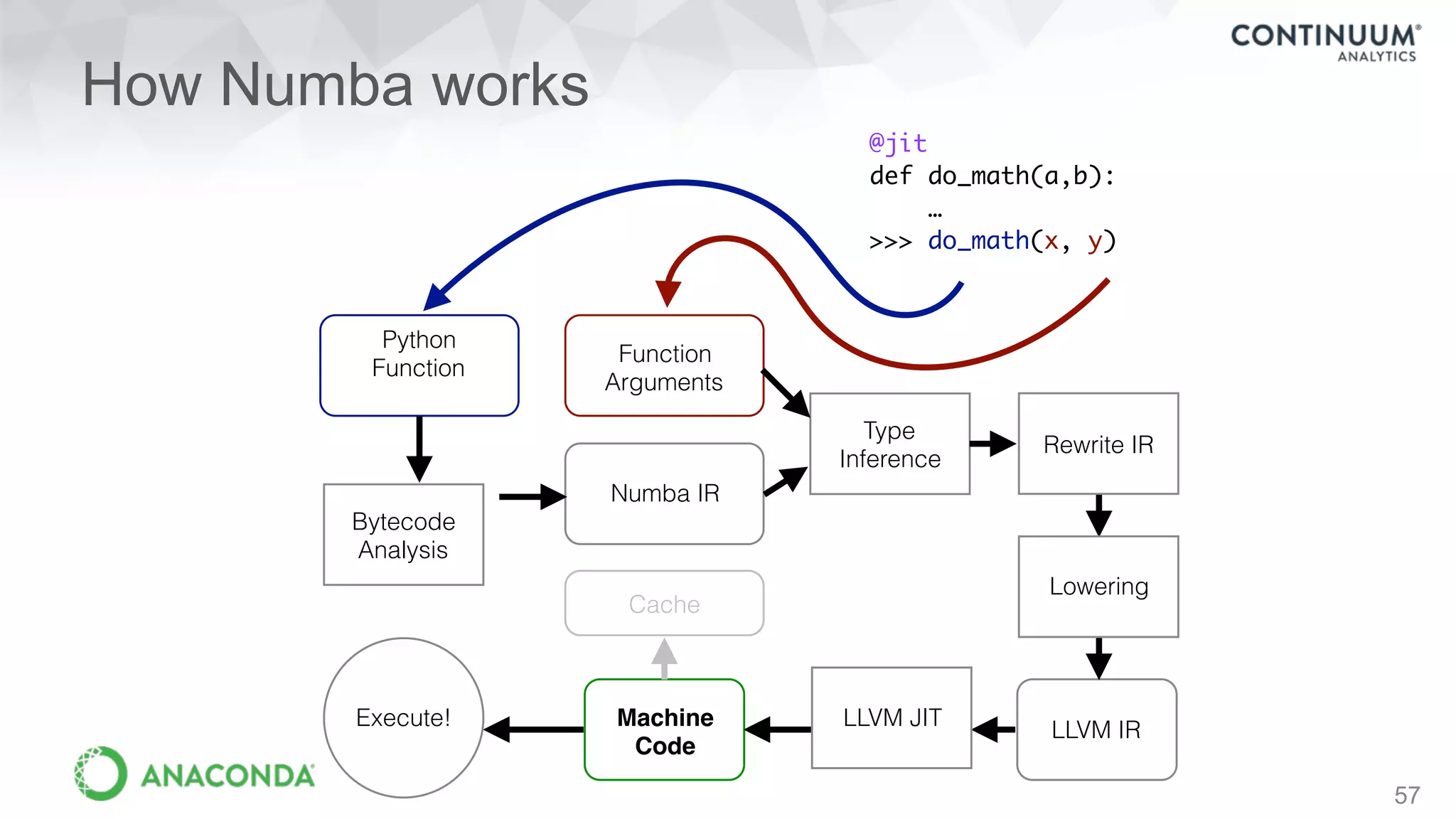
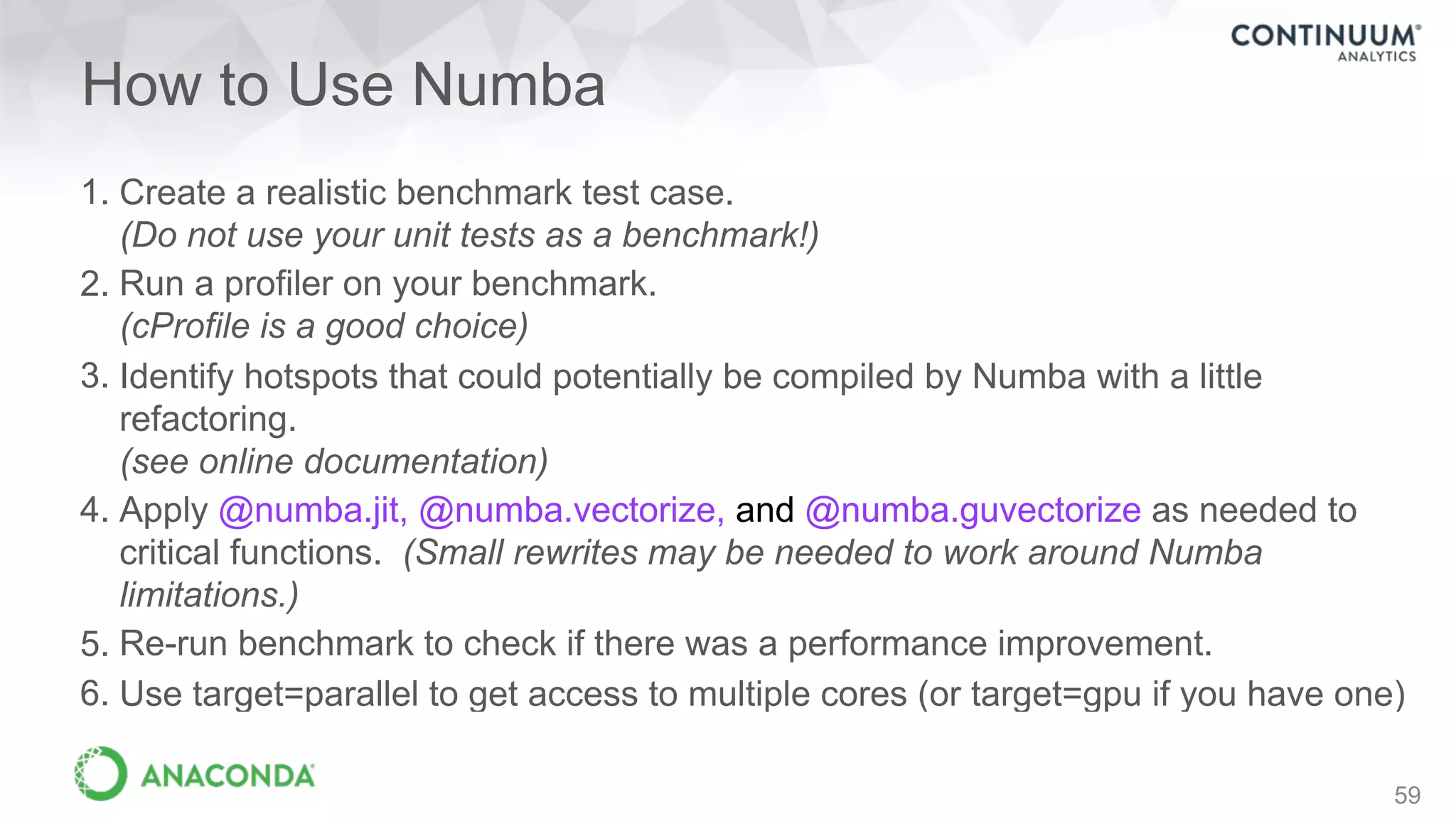
This document discusses tools for making NumPy and Pandas code faster and able to run in parallel. It introduces the Dask library, which allows users to work with large datasets in a familiar Pandas/NumPy style through parallel computing. Dask implements parallel DataFrames, Arrays, and other collections that mimic their Pandas/NumPy counterparts. It can scale computations across multiple cores on a single machine or across many machines in a cluster. The document provides examples of using Dask to analyze large CSV and text data in parallel through DataFrames and Bags. It also discusses scaling computations from a single laptop to large clusters.








![23
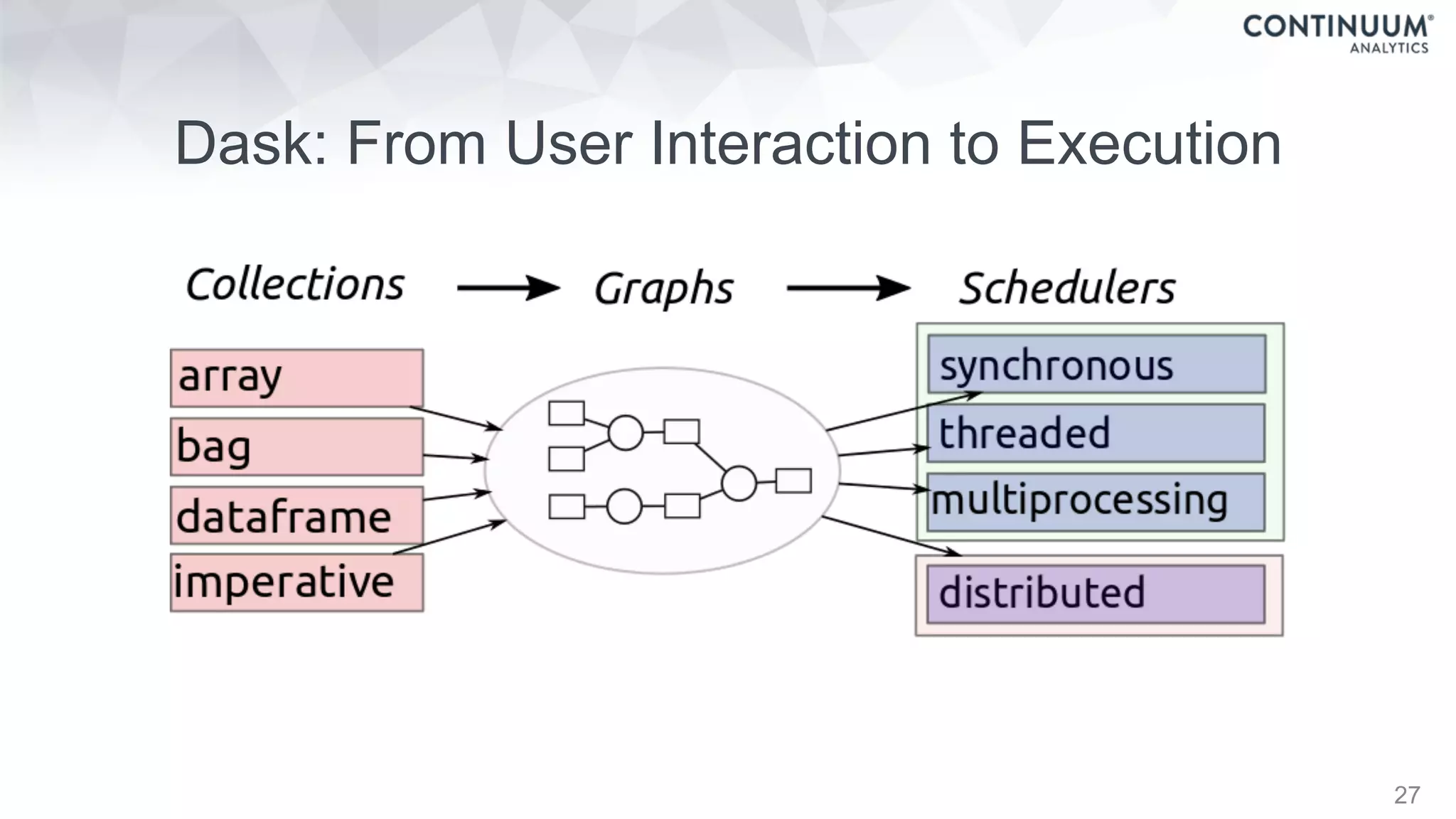
Dask Dataframes
Dask
>>> import pandas as pd
>>> df = pd.read_csv('iris.csv')
>>> df.head()
sepal_length sepal_width petal_length petal_width
species
0 5.1 3.5 1.4 0.2
Iris-setosa
1 4.9 3.0 1.4 0.2
Iris-setosa
2 4.7 3.2 1.3 0.2
Iris-setosa
3 4.6 3.1 1.5 0.2
Iris-setosa
4 5.0 3.6 1.4 0.2
Iris-setosa
>>> max_sepal_length_setosa = df[df.species ==
'setosa'].sepal_length.max()
5.7999999999999998
>>> import dask.dataframe as dd
>>> ddf = dd.read_csv('*.csv')
>>> ddf.head()
sepal_length sepal_width petal_length petal_width
species
0 5.1 3.5 1.4 0.2
Iris-setosa
1 4.9 3.0 1.4 0.2
Iris-setosa
2 4.7 3.2 1.3 0.2
Iris-setosa
3 4.6 3.1 1.5 0.2
Iris-setosa
4 5.0 3.6 1.4 0.2
Iris-setosa
…
>>> d_max_sepal_length_setosa = ddf[ddf.species ==
'setosa'].sepal_length.max()
>>> d_max_sepal_length_setosa.compute()
5.7999999999999998](https://image.slidesharecdn.com/fastandscalablepython-160622041030/75/Fast-and-Scalable-Python-23-2048.jpg)
![24
Dask Arrays
>>> import numpy as np
>>> np_ones = np.ones((5000, 1000))
>>> np_ones
array([[ 1., 1., 1., ..., 1., 1., 1.],
[ 1., 1., 1., ..., 1., 1., 1.],
[ 1., 1., 1., ..., 1., 1., 1.],
...,
[ 1., 1., 1., ..., 1., 1., 1.],
[ 1., 1., 1., ..., 1., 1., 1.],
[ 1., 1., 1., ..., 1., 1., 1.]])
>>> np_y = np.log(np_ones + 1)[:5].sum(axis=1)
>>> np_y
array([ 693.14718056, 693.14718056, 693.14718056,
693.14718056, 693.14718056])
>>> import dask.array as da
>>> da_ones = da.ones((5000000, 1000000),
chunks=(1000, 1000))
>>> da_ones.compute()
array([[ 1., 1., 1., ..., 1., 1., 1.],
[ 1., 1., 1., ..., 1., 1., 1.],
[ 1., 1., 1., ..., 1., 1., 1.],
...,
[ 1., 1., 1., ..., 1., 1., 1.],
[ 1., 1., 1., ..., 1., 1., 1.],
[ 1., 1., 1., ..., 1., 1., 1.]])
>>> da_y = da.log(da_ones + 1)[:5].sum(axis=1)
>>> np_da_y = np.array(da_y) #fits in memory
array([ 693.14718056, 693.14718056, 693.14718056,
693.14718056, …, 693.14718056])
# Result doesn’t fit in memory
>>> da_y.to_hdf5('myfile.hdf5', 'result')
Dask](https://image.slidesharecdn.com/fastandscalablepython-160622041030/75/Fast-and-Scalable-Python-24-2048.jpg)
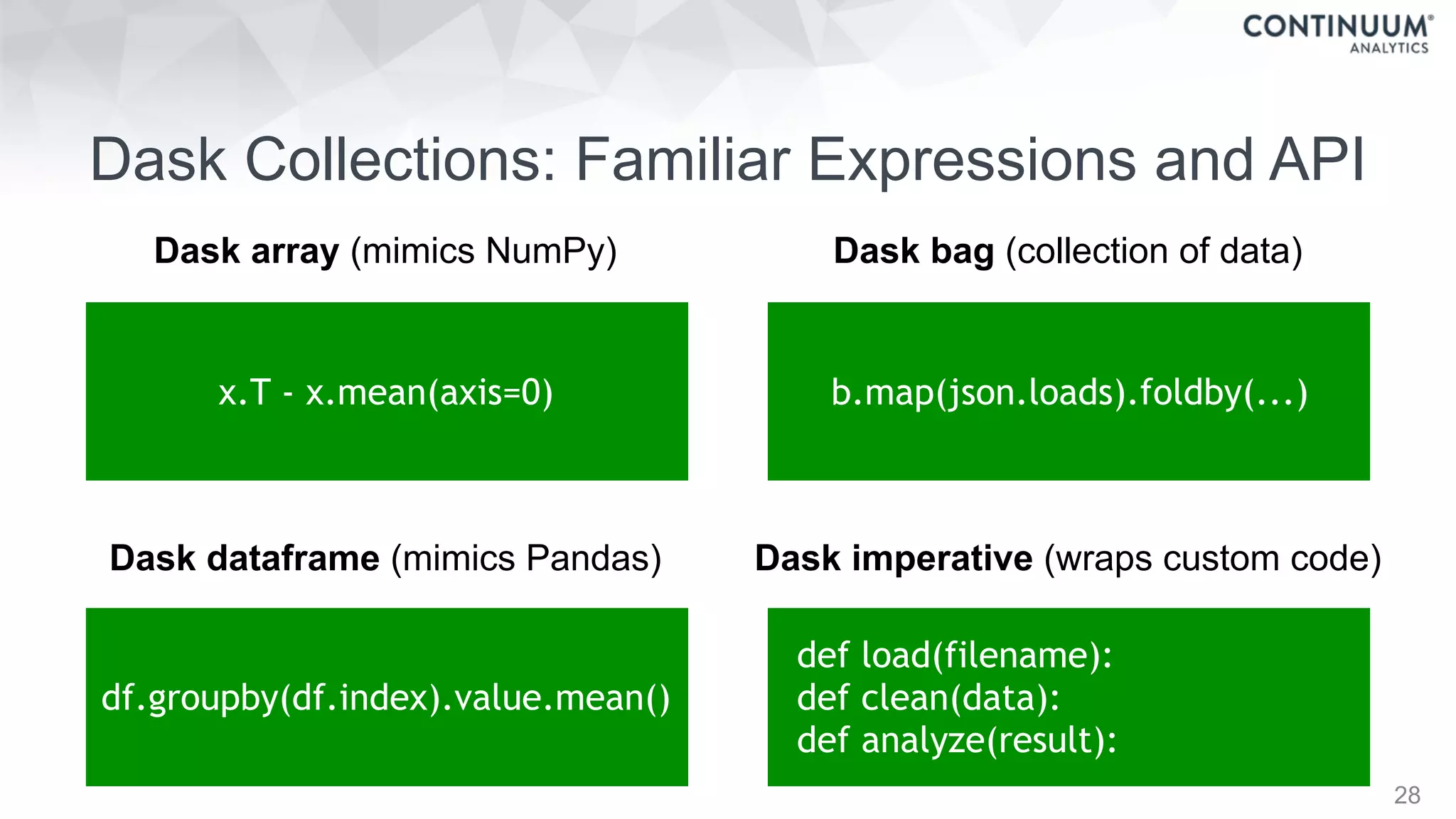
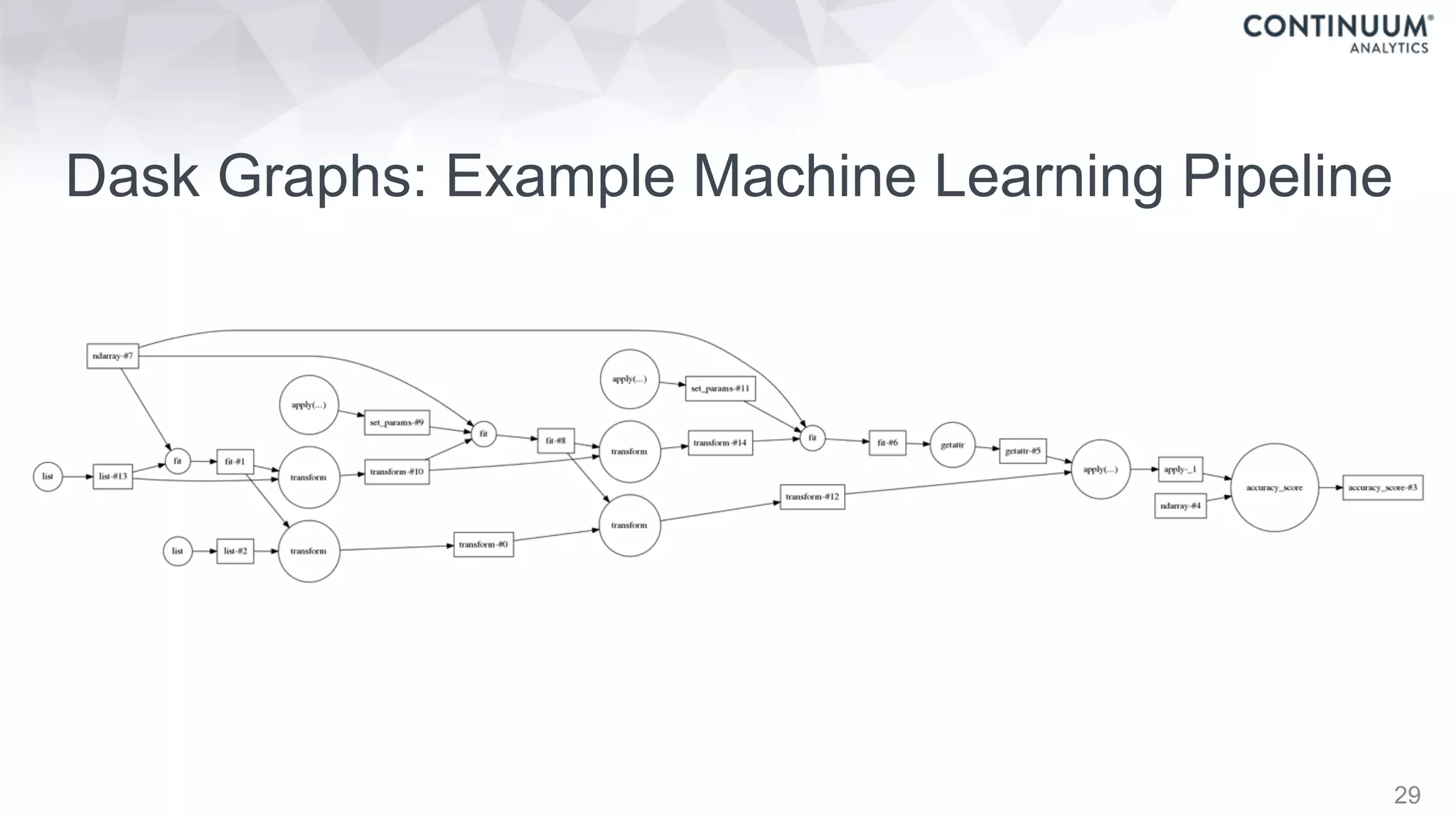
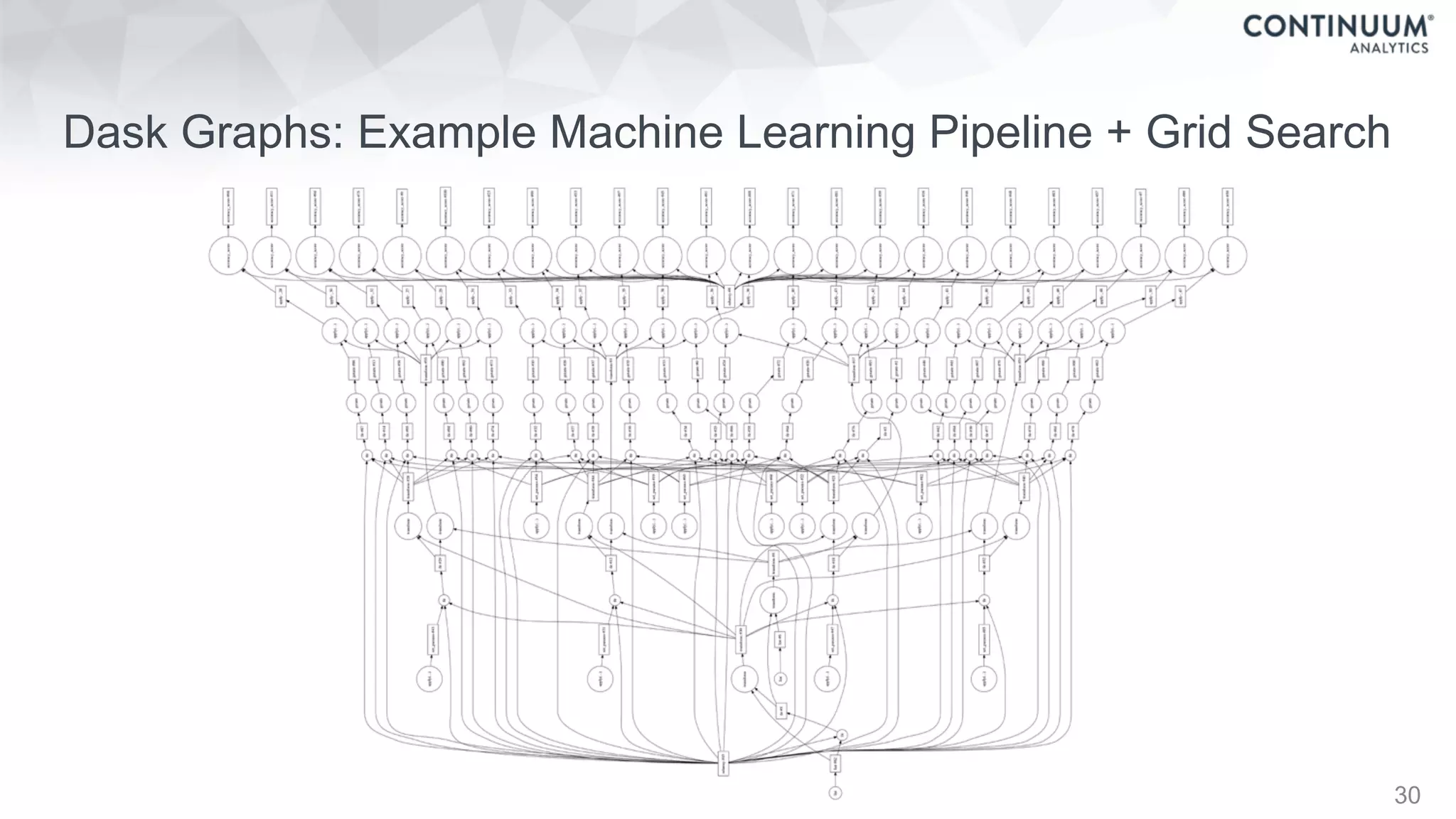
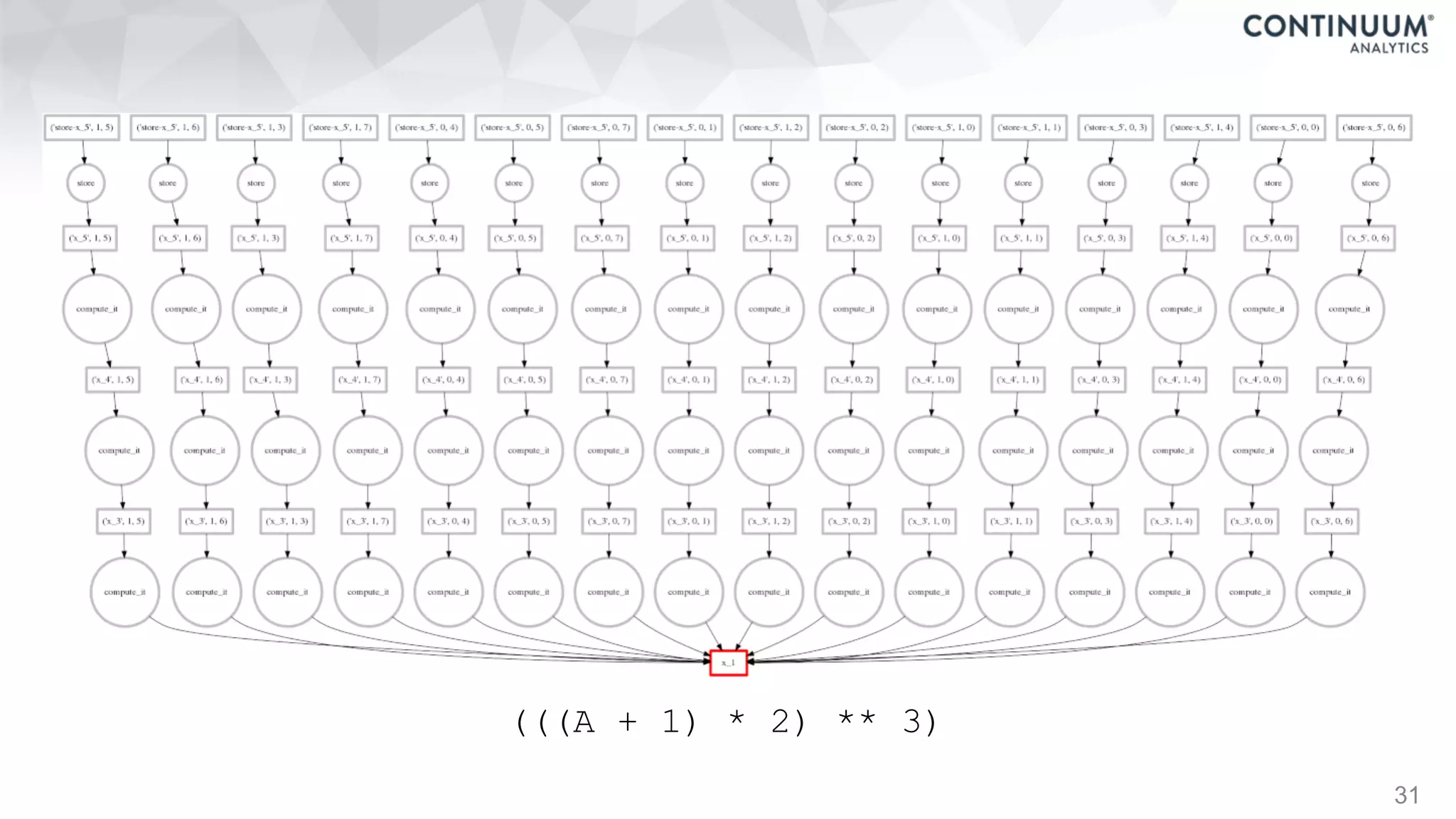

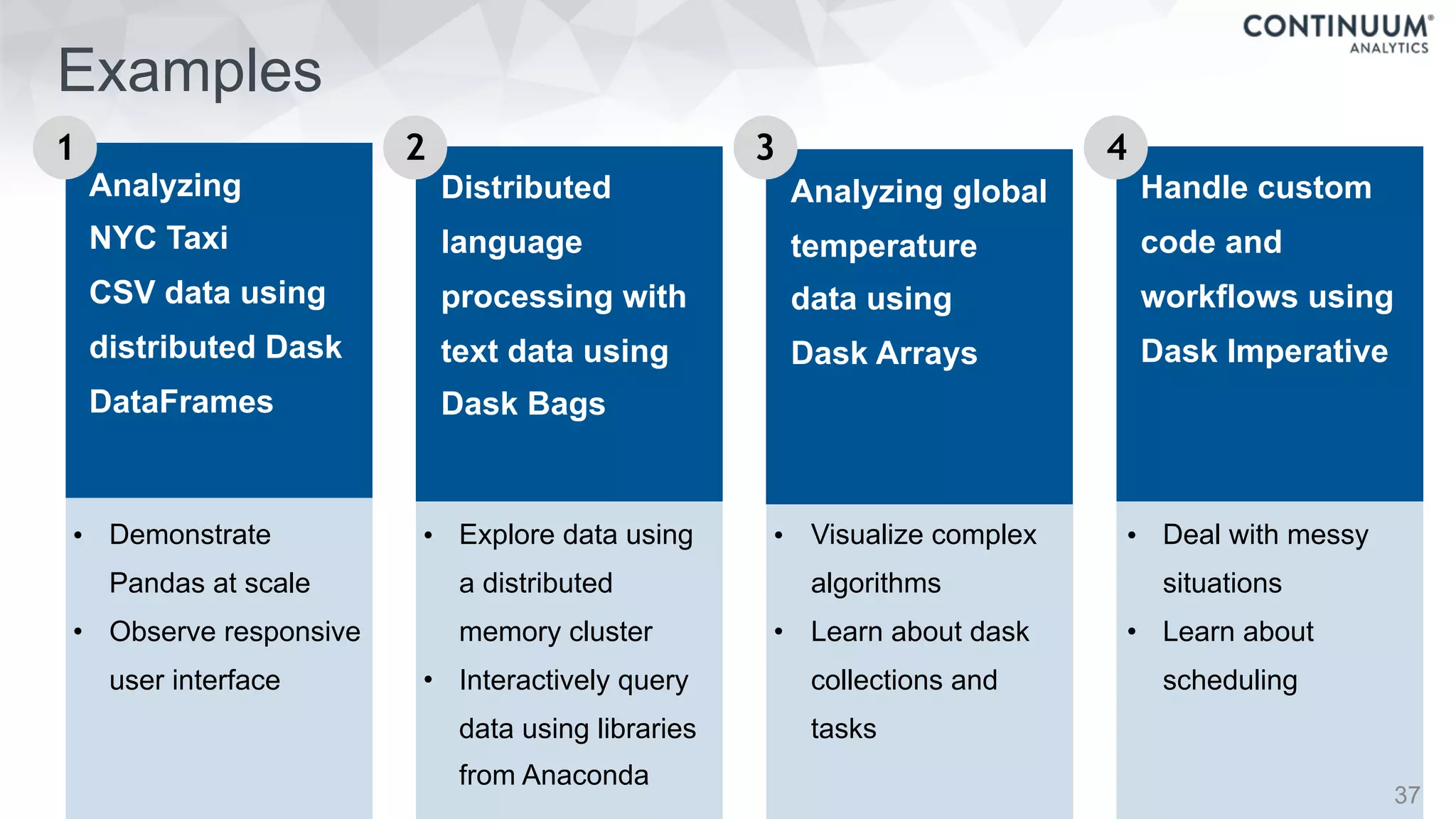
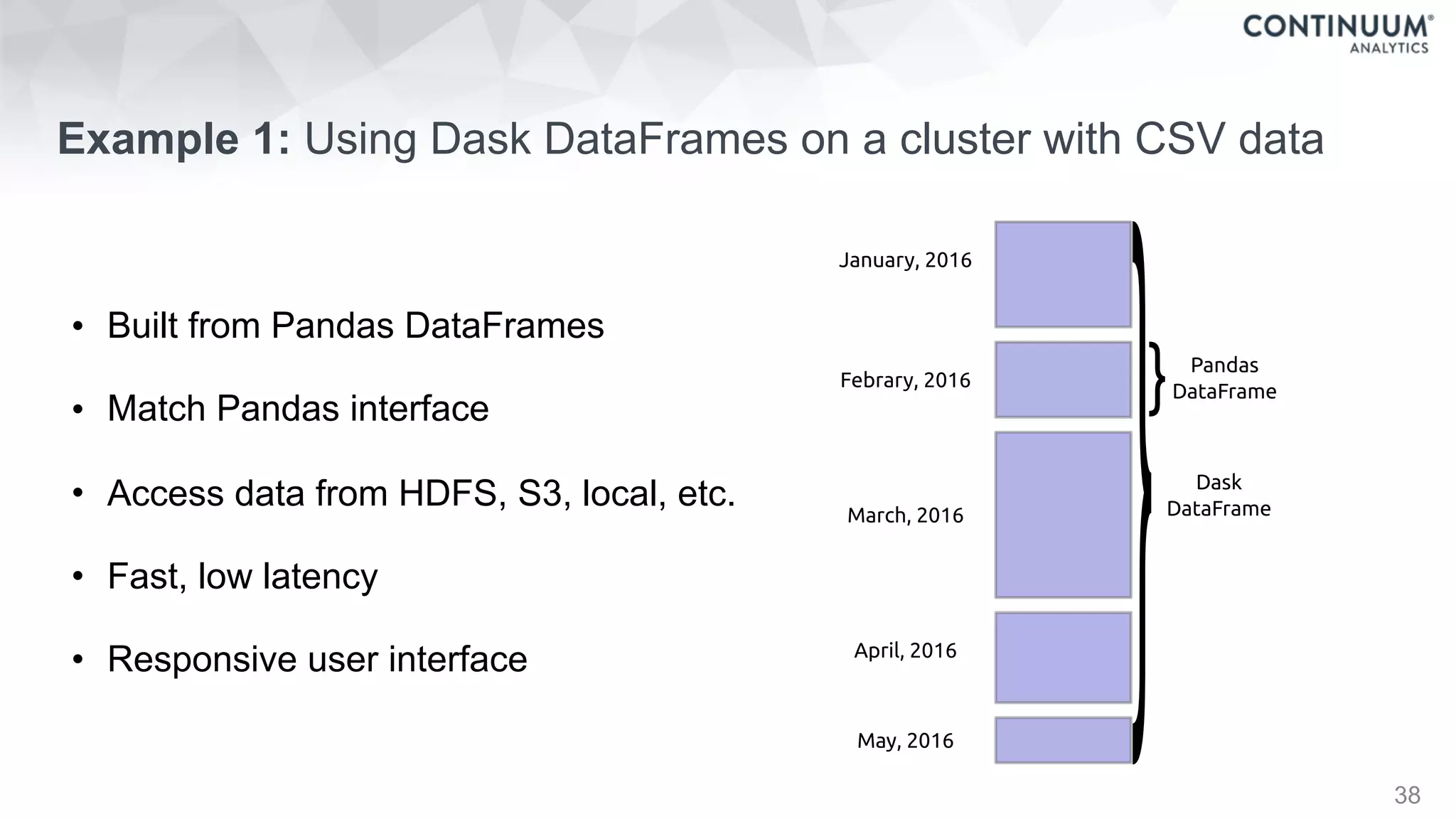
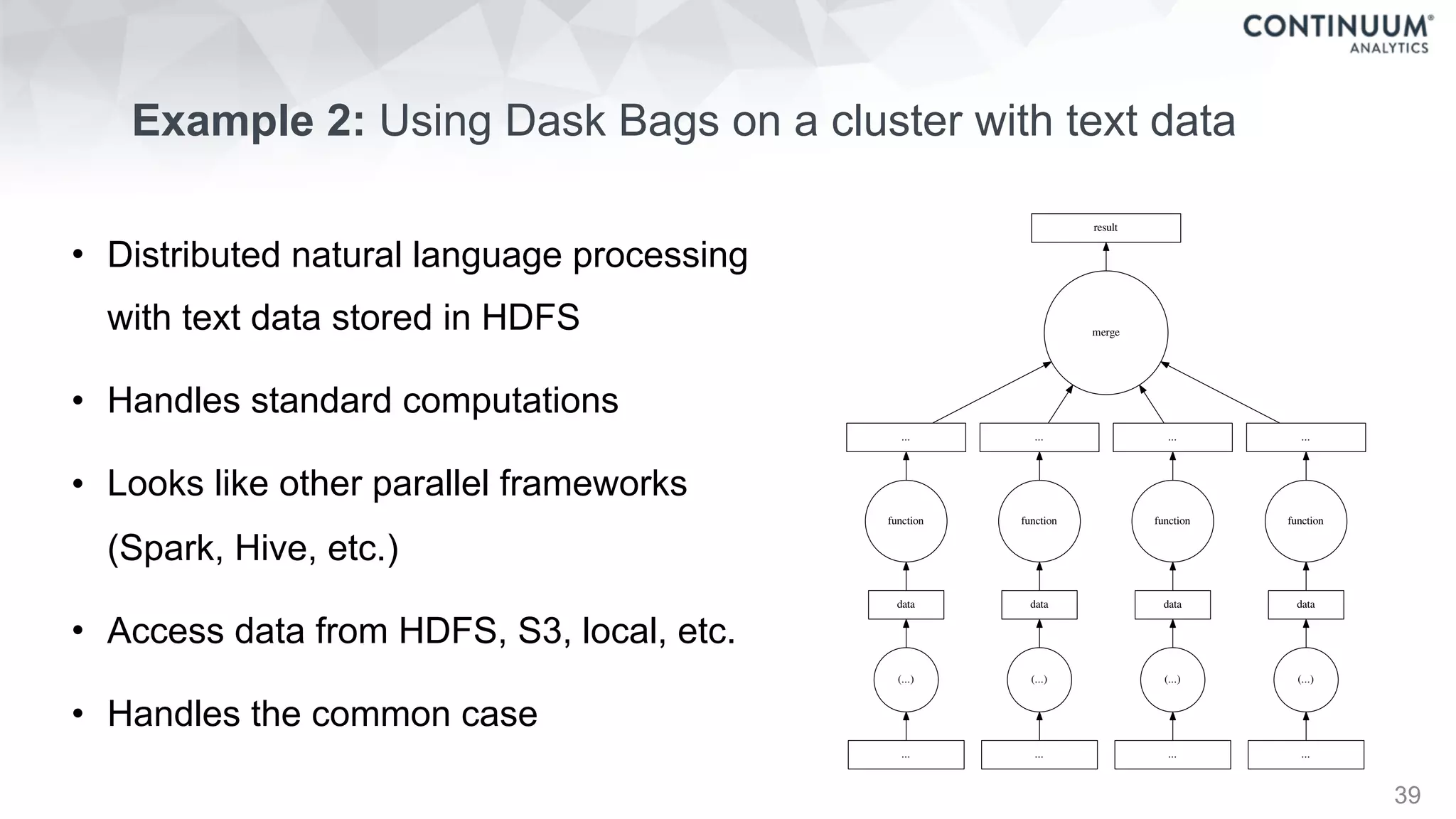
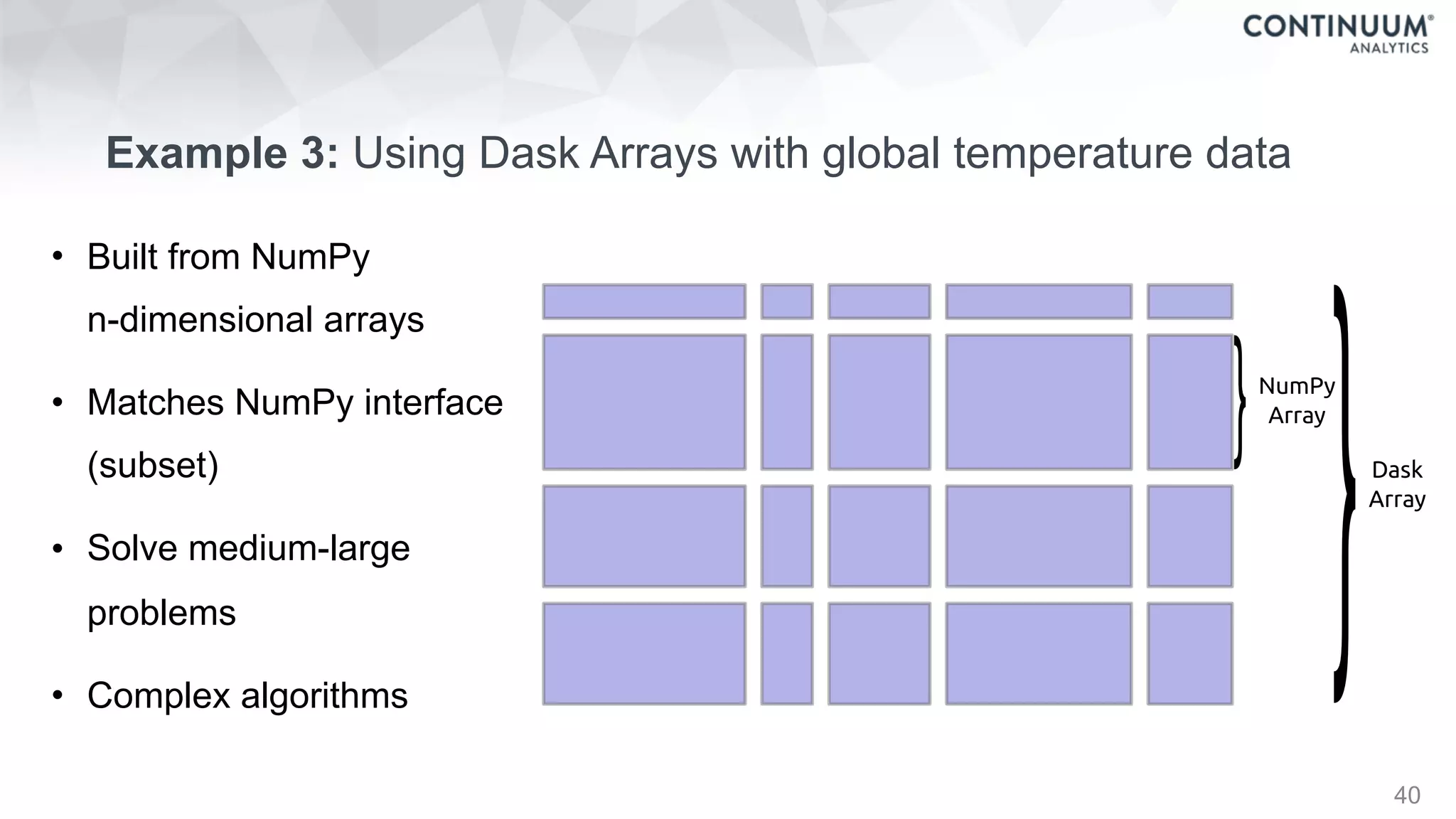
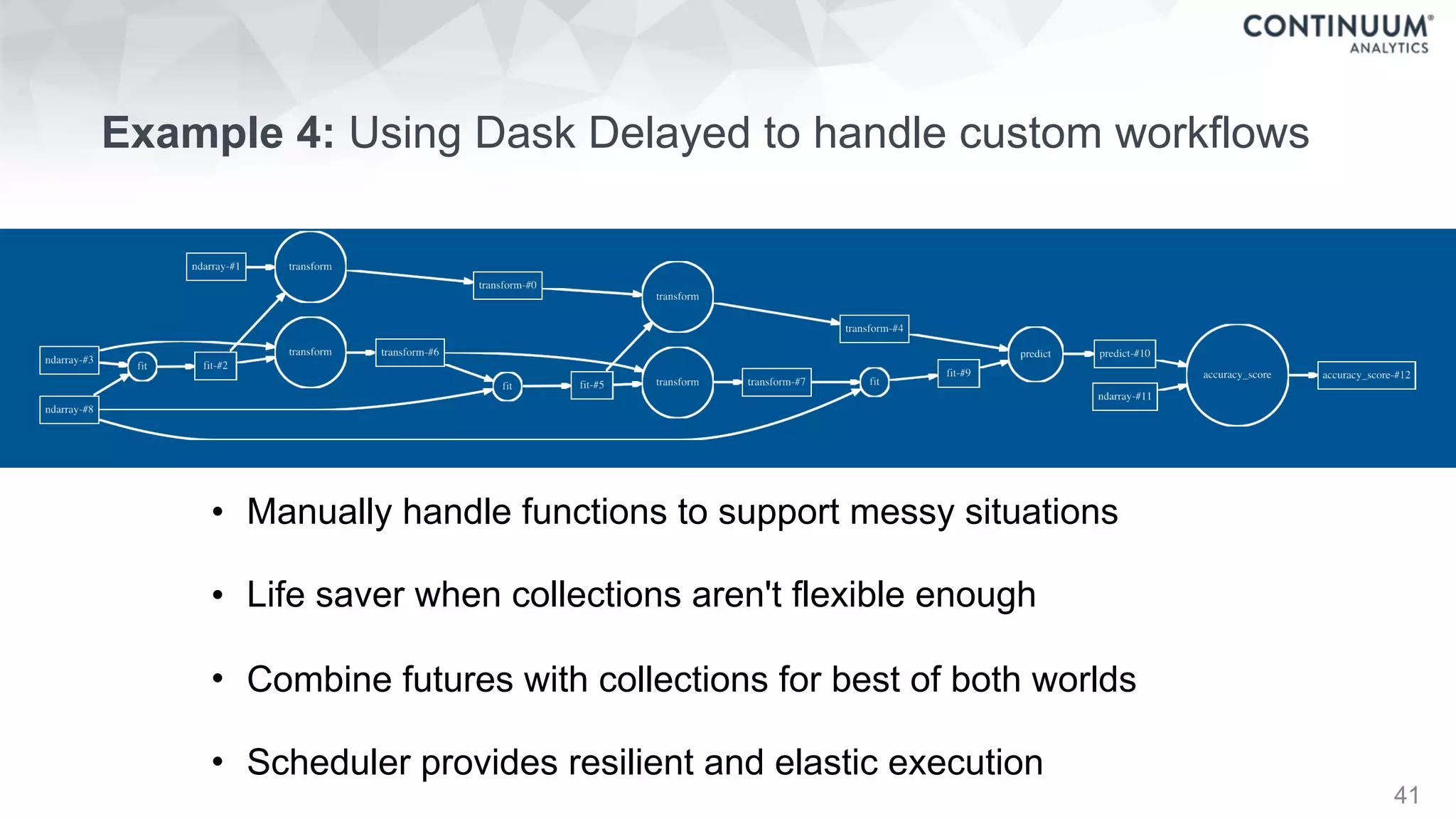

![44
+ - / * ^ []
join, groupby, filter
map, sort, take
where, topk
datashape,dtype,
shape,stride
hdf5,json,csv,xls
protobuf,avro,...
NumPy,Pandas,R,
Julia,K,SQL,Spark,
Mongo,Cassandra,...](https://image.slidesharecdn.com/fastandscalablepython-160622041030/75/Fast-and-Scalable-Python-44-2048.jpg)

![Blaze
47
iris[['sepal_length', 'species']]Select columns
log(iris.sepal_length * 10)Operate
Reduce iris.sepal_length.mean()
Split-apply
-combine
by(iris.species, shortest=iris.petal_length.min(),
longest=iris.petal_length.max(),
average=iris.petal_length.mean())
Add new
columns
transform(iris, sepal_ratio = iris.sepal_length /
iris.sepal_width, petal_ratio = iris.petal_length /
iris.petal_width)
Text matching iris.like(species='*versicolor')
iris.relabel(petal_length='PETAL-LENGTH',
petal_width='PETAL-WIDTH')
Relabel columns
Filter iris[(iris.species == 'Iris-setosa') & (iris.sepal_length > 5.0)]](https://image.slidesharecdn.com/fastandscalablepython-160622041030/75/Fast-and-Scalable-Python-47-2048.jpg)

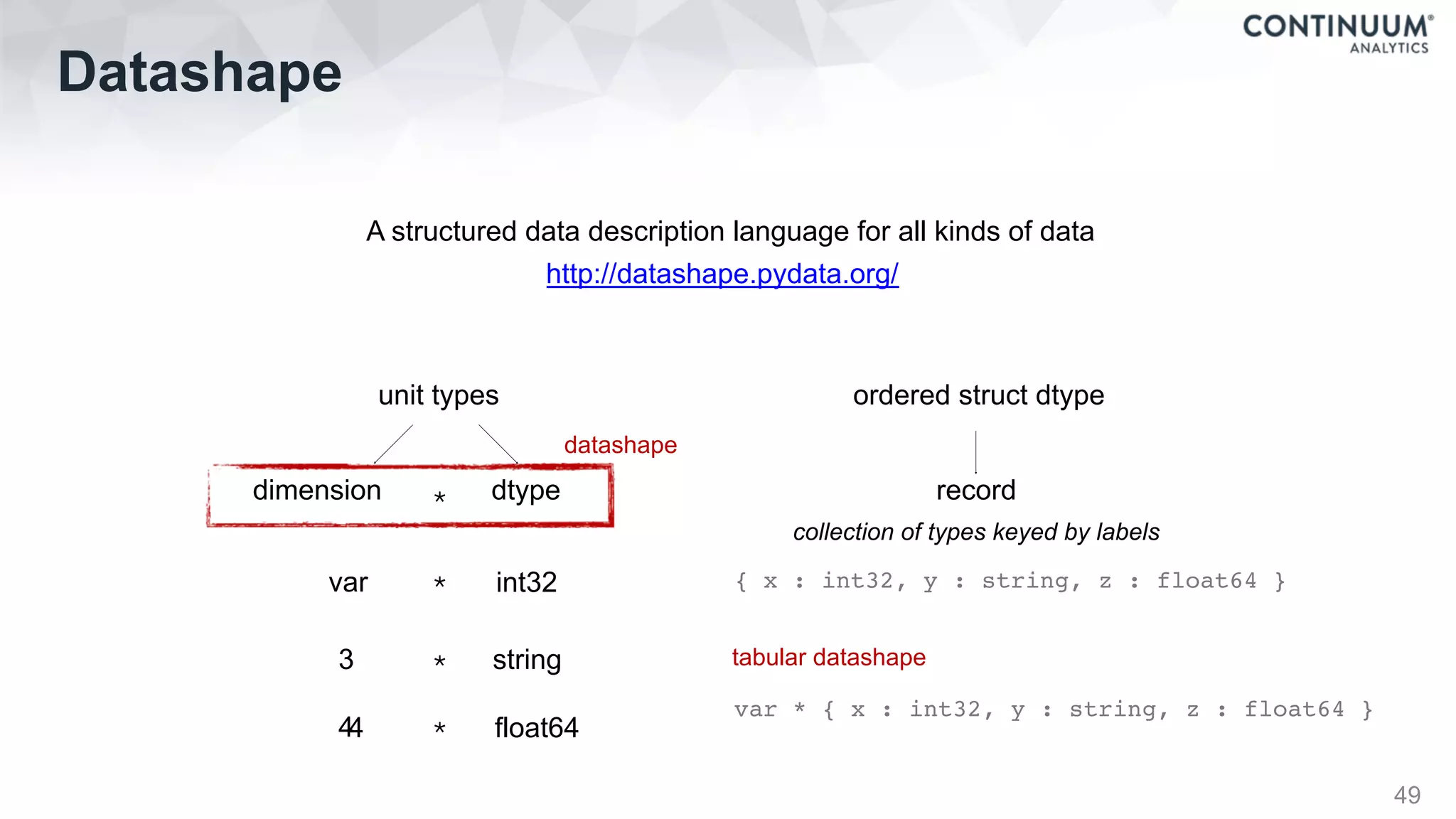
![Datashape
50
{
flowersdb: {
iris: var * {
petal_length: float64,
petal_width: float64,
sepal_length: float64,
sepal_width: float64,
species: string
}
},
iriscsv: var * {
sepal_length: ?float64,
sepal_width: ?float64,
petal_length: ?float64,
petal_width: ?float64,
species: ?string
},
irisjson: var * {
petal_length: float64,
petal_width: float64,
sepal_length: float64,
sepal_width: float64,
species: string
},
irismongo: 150 * {
petal_length: float64,
petal_width: float64,
sepal_length: float64,
sepal_width: float64,
species: string
}
}
# Arrays of Structures
100 * {
name: string,
birthday: date,
address: {
street: string,
city: string,
postalcode: string,
country: string
}
}
# Structure of Arrays
{
x: 100 * 100 * float32,
y: 100 * 100 * float32,
u: 100 * 100 * float32,
v: 100 * 100 * float32,
}
# Function prototype
(3 * int32, float64) -> 3 * float64
# Function prototype with broadcasting dimensions
(A... * int32, A... * int32) -> A... * int32
# Arrays
3 * 4 * int32
3 * 4 * int32
10 * var * float64
3 * complex[float64]](https://image.slidesharecdn.com/fastandscalablepython-160622041030/75/Fast-and-Scalable-Python-50-2048.jpg)

![Blaze Client
52
>>> from blaze import Data
>>> s = Data('blaze://localhost:6363')
>>> t.fields
[u'iriscsv', u'irisdb', u'irisjson', u’irismongo']
>>> t.iriscsv
sepal_length sepal_width petal_length petal_width species
0 5.1 3.5 1.4 0.2 Iris-setosa
1 4.9 3.0 1.4 0.2 Iris-setosa
2 4.7 3.2 1.3 0.2 Iris-setosa
>>> t.irisdb
petal_length petal_width sepal_length sepal_width species
0 1.4 0.2 5.1 3.5 Iris-setosa
1 1.4 0.2 4.9 3.0 Iris-setosa
2 1.3 0.2 4.7 3.2 Iris-setosa](https://image.slidesharecdn.com/fastandscalablepython-160622041030/75/Fast-and-Scalable-Python-52-2048.jpg)




![Generalized Ufunc
64
@guvectorize(‘f8[:], f8[:], f8[:]’, ‘(n),(n)->()’)
def dot2(a,b,c):
c[0]=a[0]*b[0] + a[1]*b[1]
>>> a, b = np.random.randn(10000,2), np.random.randn(10000,2)
>>> z1 = np.einsum(‘ij,ij->i’, a, b)
>>> z2 = dot2(a, b) # uses last dimension as in each kernel
This can create quite a bit of computation with very little code.
Numba creates a fast machine-code kernel from the Python template
and calls it for every element in the arrays.
3.8x faster](https://image.slidesharecdn.com/fastandscalablepython-160622041030/75/Fast-and-Scalable-Python-64-2048.jpg)