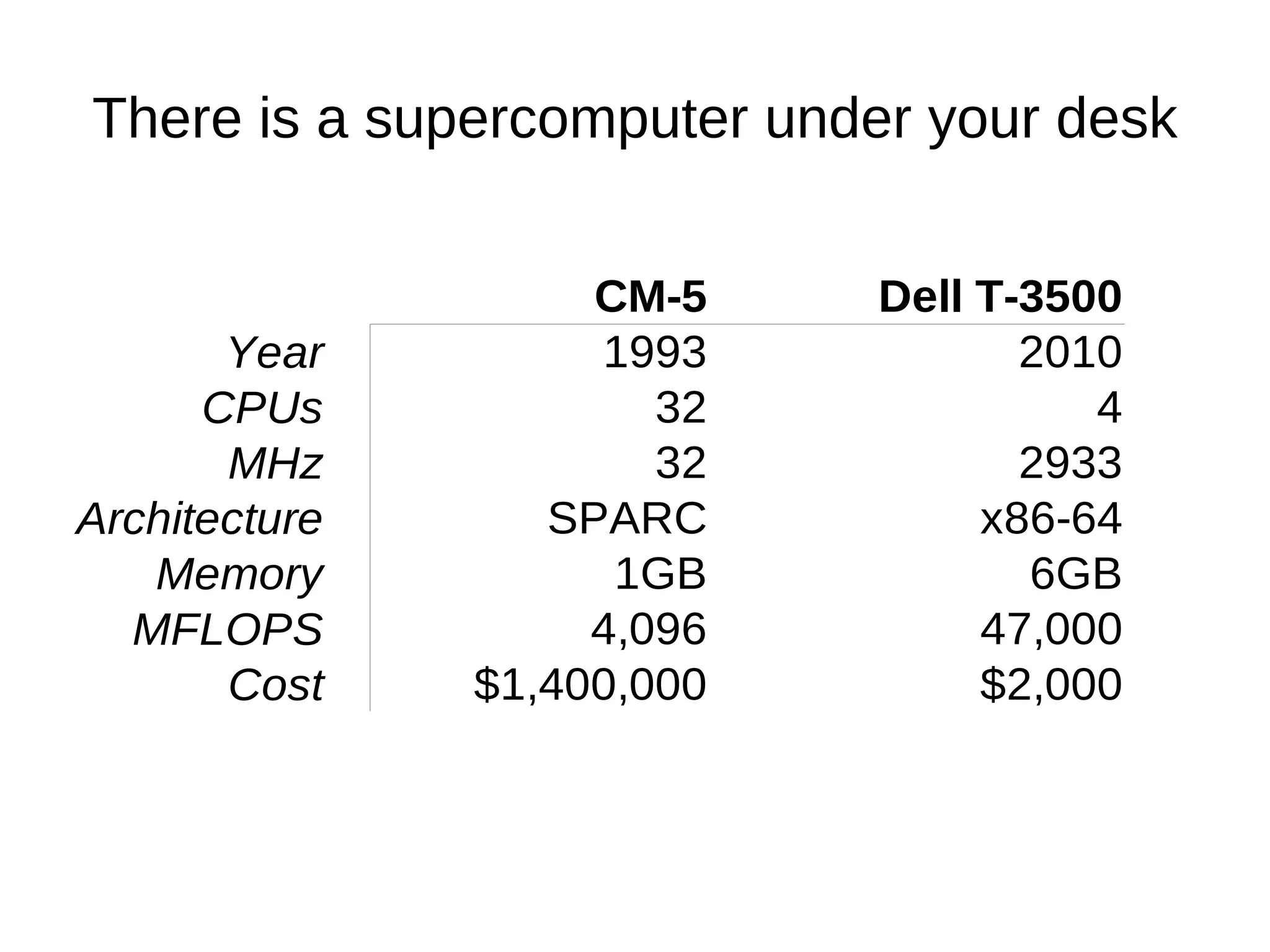
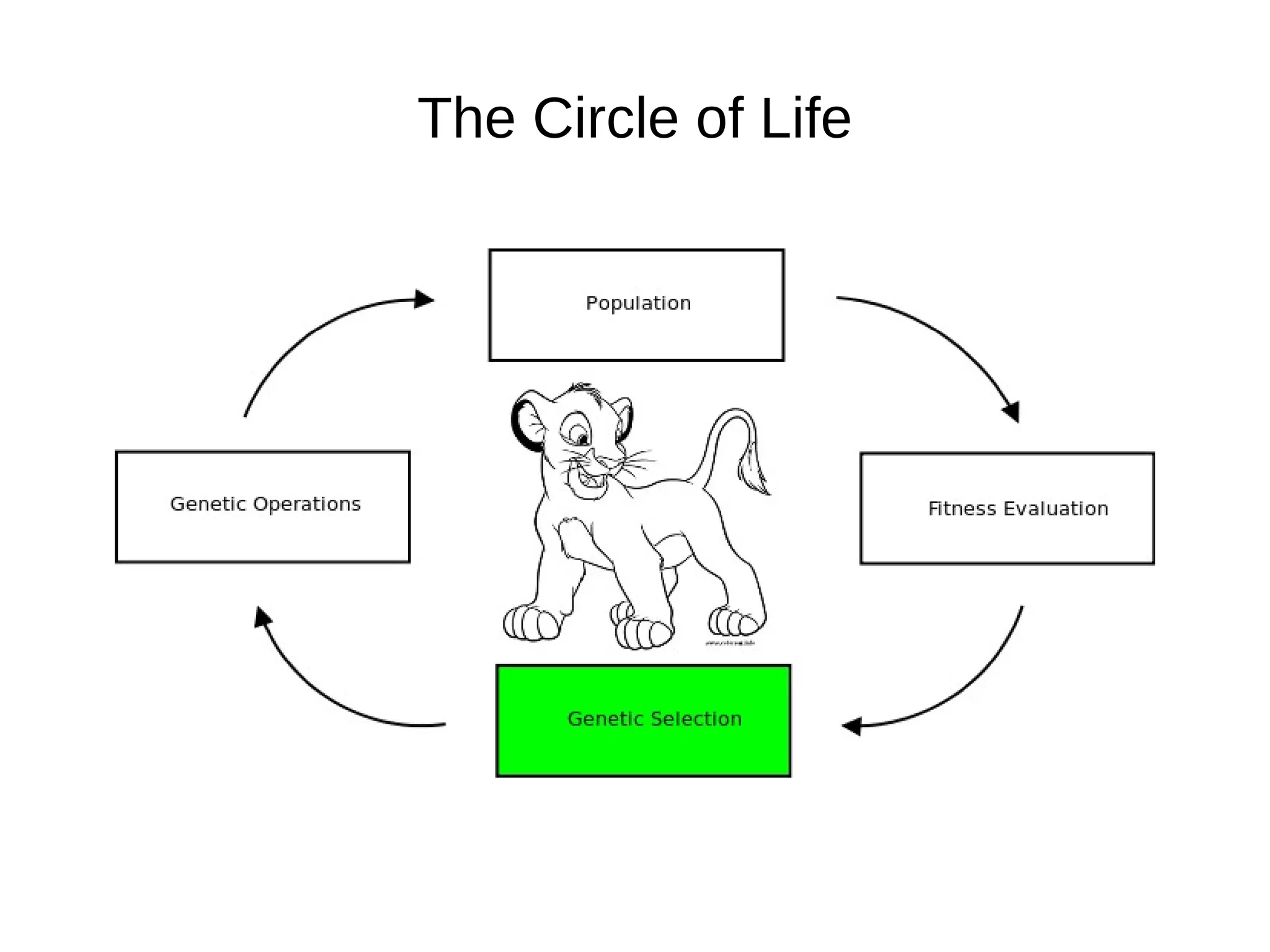
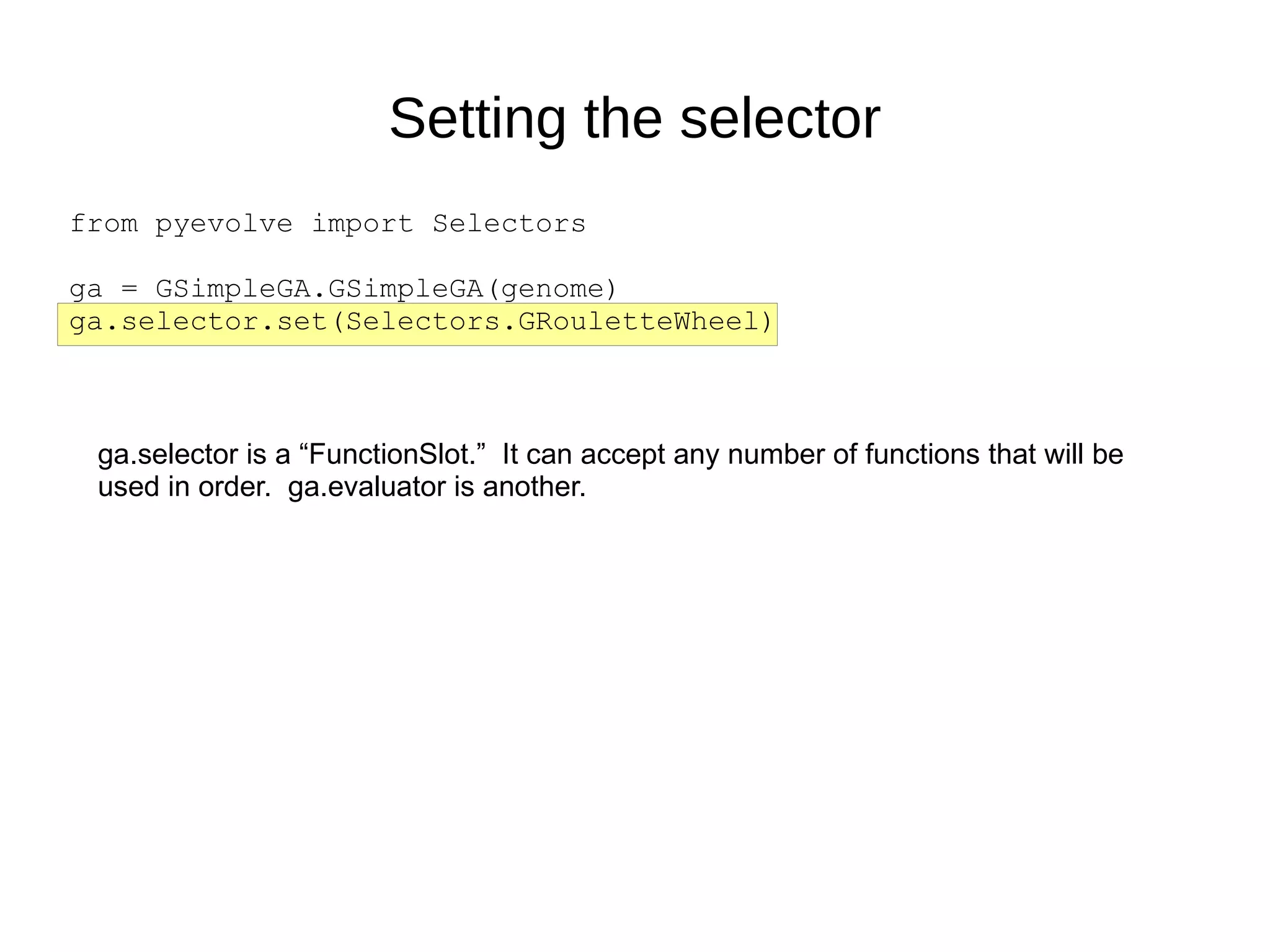
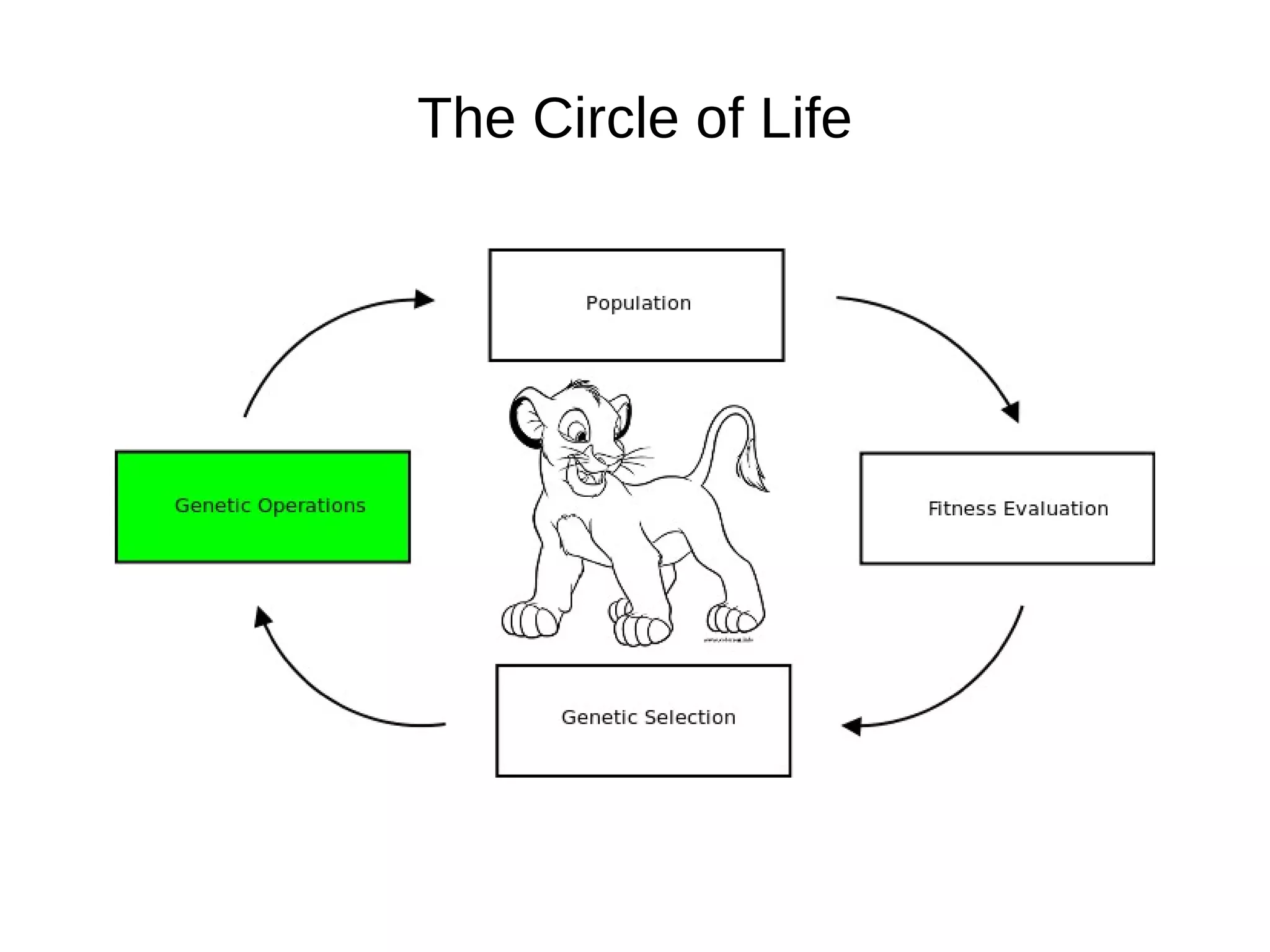
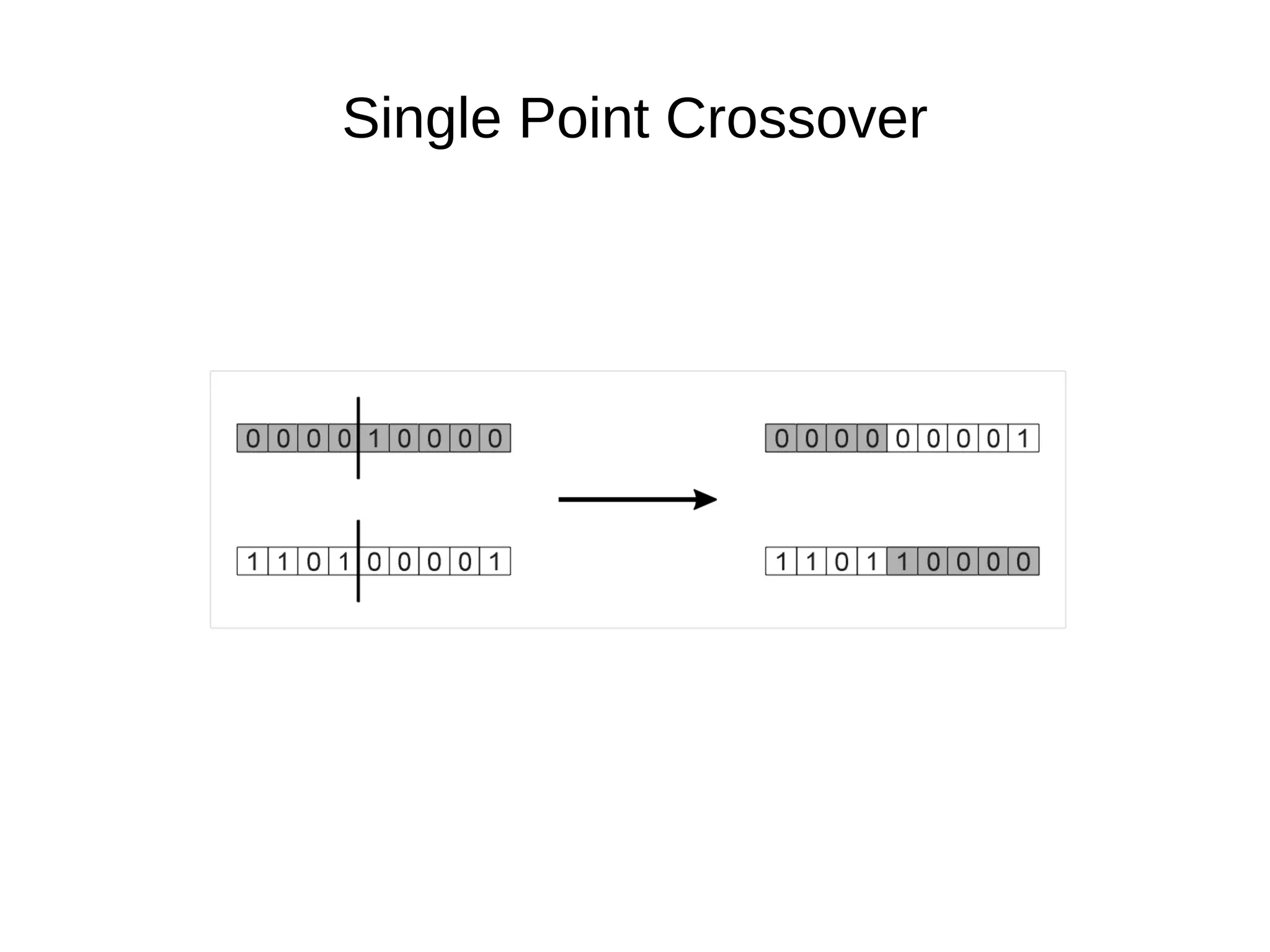
The document discusses genetic algorithms and genetic programming in Python. It describes how genetic algorithms are inspired by natural selection and genetics, using techniques like selection, crossover, and mutation to evolve solutions to problems. It provides examples of using the Python library PyEvolve to implement genetic algorithms and genetic programming to solve problems like minimizing test functions and forecasting temperatures.


















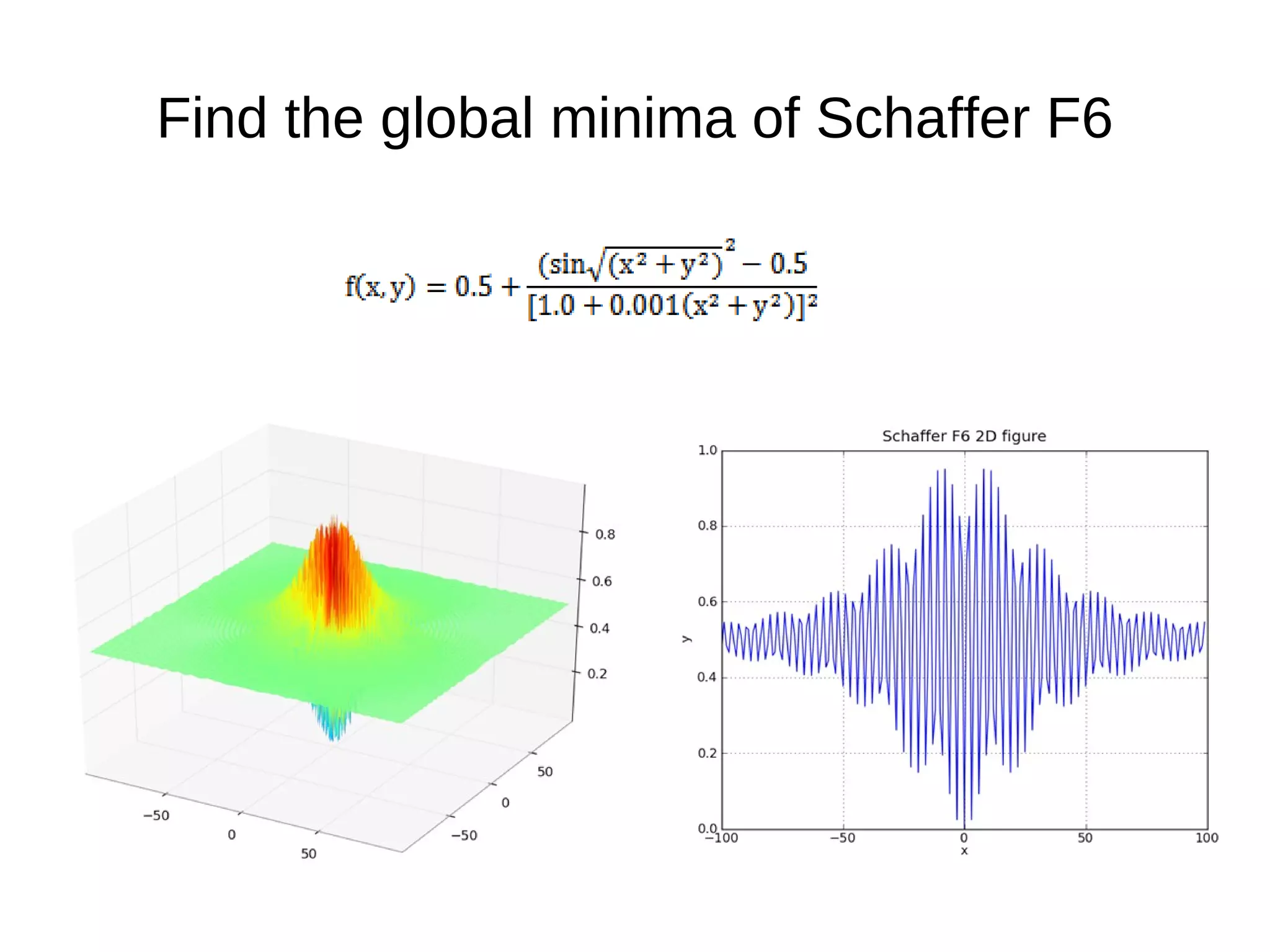










![The Fitness Function def schafferF6(genome): x2y2 = genome[0]**2 + genome[1]**2 t1 = math.sin(math.sqrt(x2y2)); t2 = 1.0 + 0.001*(x2y2); score = 0.5 + (t1**2 - 0.5)/(t2*t2) return score genome = G1DList.G1DList(2) genome.setParams(rangemin=-100.0, rangemax=100.0, bestrawscore=0.0000, rounddecimal=4) genome.initializator.set(Initializators.G1DListInitializatorReal) genome.evaluator.set(schafferF6)](https://image.slidesharecdn.com/presentation-100811152315-phpapp02/75/Genetic-Programming-in-Python-45-2048.jpg)

![The GA Engine from pyevolve import GSimpleGA … create genome … ga = GSimpleGA.GSimpleGA(genome, seed=123) ga.setMinimax(Consts.minimaxType["minimize"]) ga.evolve(freq_stats=1000) print ga.bestIndividual() Output: Gen. 0 (0.00%): Max/Min/Avg Fitness(Raw) [0.60(0.82)/0.37(0.09)/0.50(0.50)] Gen. 1000 (12.50%): Max/Min/Avg Fitness(Raw) [0.30(0.97)/0.23(0.01)/0.25(0.25)] Gen. 2000 (25.00%): Max/Min/Avg Fitness(Raw) [0.21(0.99)/0.17(0.01)/0.18(0.18)] Gen. 3000 (37.50%): Max/Min/Avg Fitness(Raw) [0.26(0.99)/0.21(0.00)/0.22(0.22)] Evolution stopped by Termination Criteria function ! Gen. 3203 (40.04%): Max/Min/Avg Fitness(Raw) [0.30(0.99)/0.23(0.00)/0.25(0.25)] Total time elapsed: 14.357 seconds. - GenomeBase Score: 0.000005 Fitness: 0.232880 - G1DList List size: 2 List: [0.0020881039453384299, 0.00043589670629584631]](https://image.slidesharecdn.com/presentation-100811152315-phpapp02/75/Genetic-Programming-in-Python-47-2048.jpg)
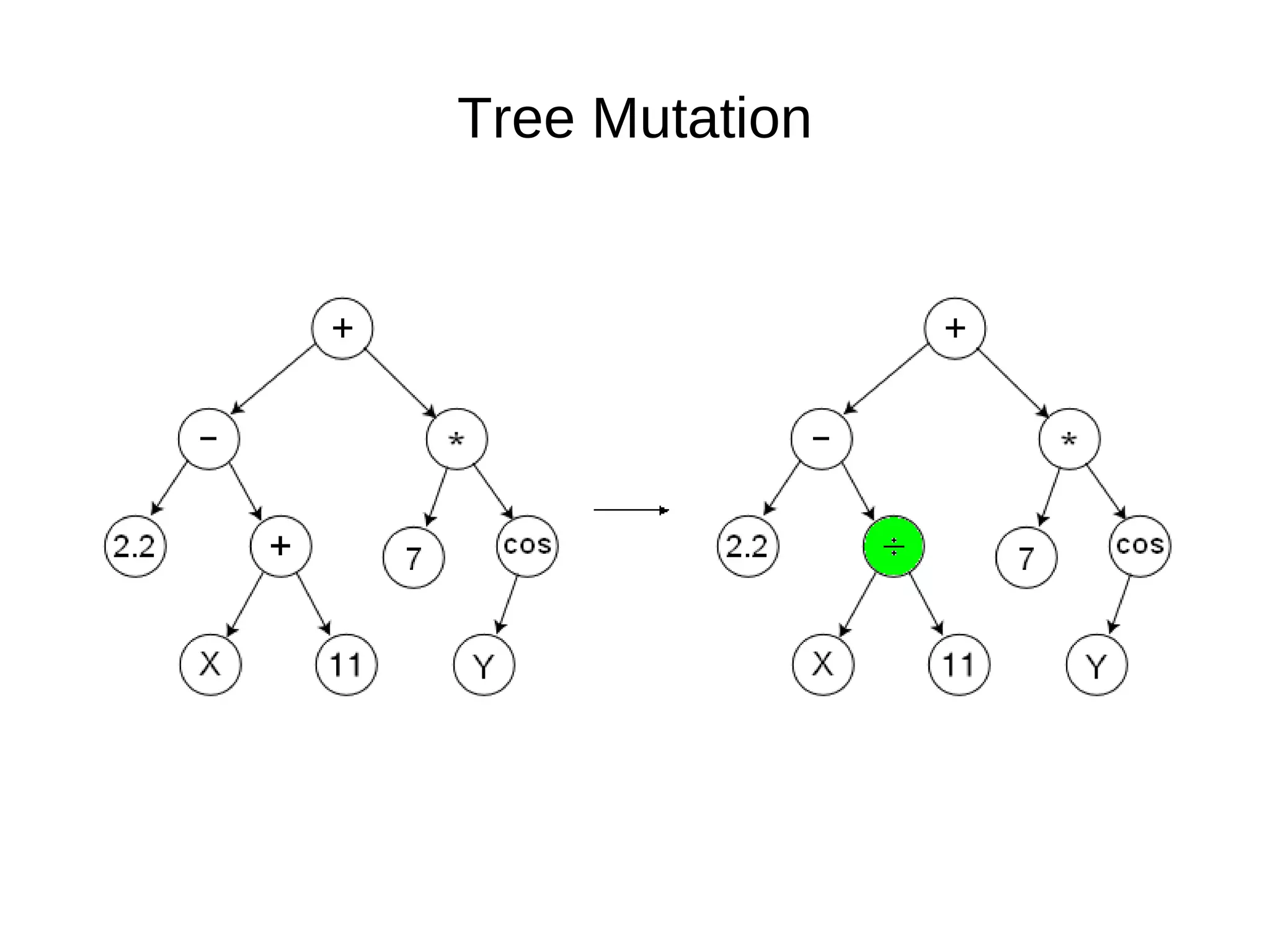





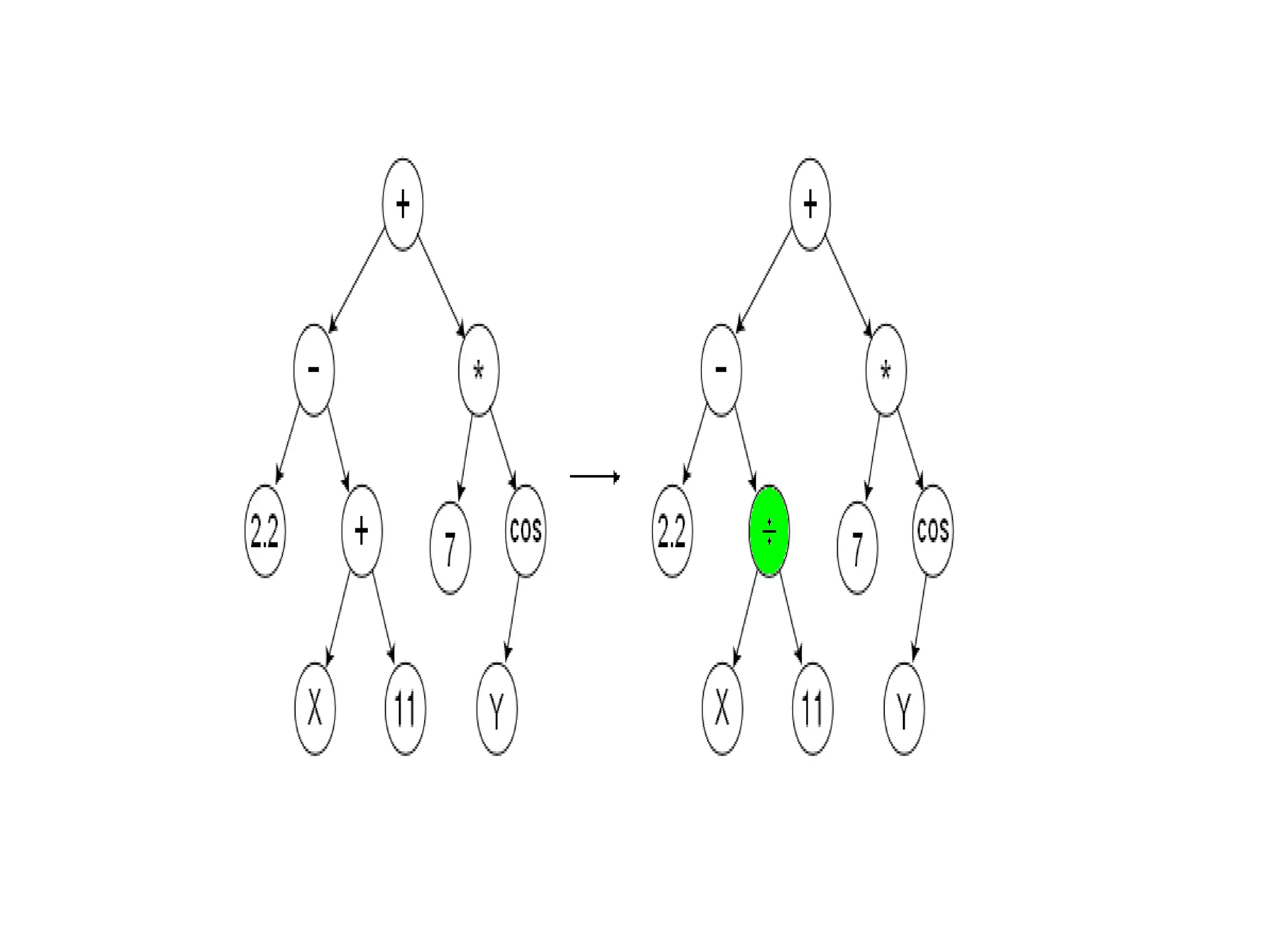
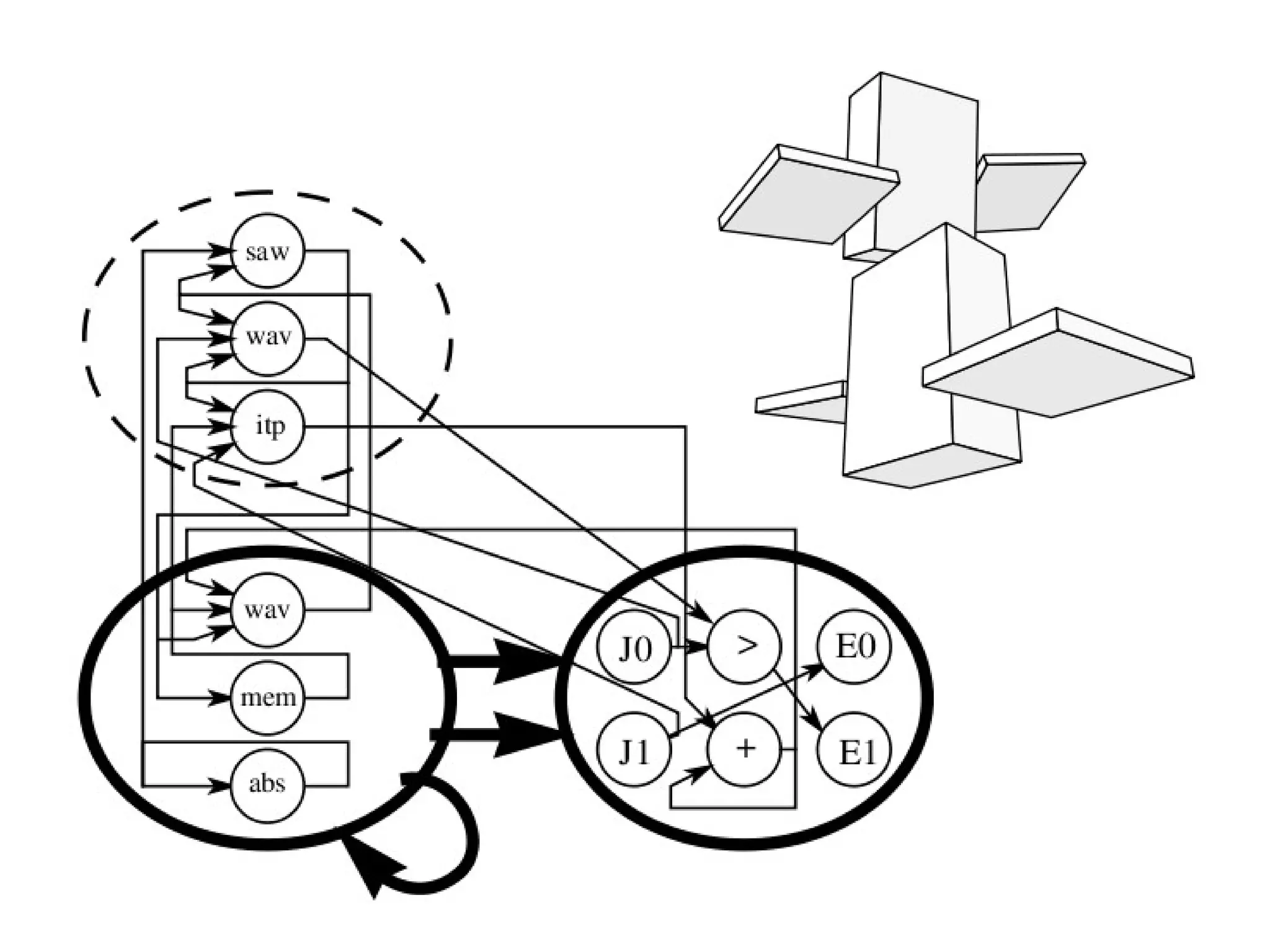



















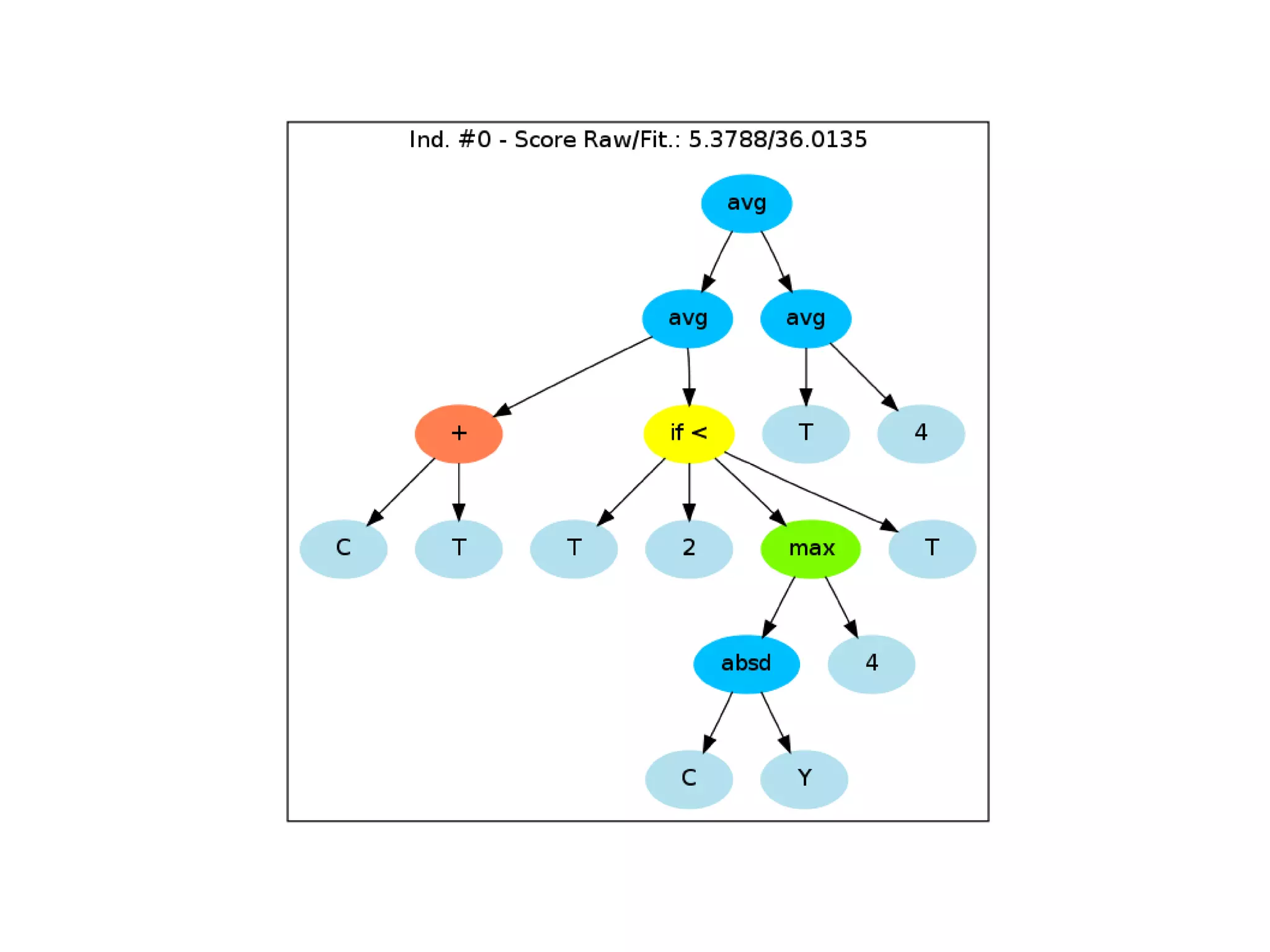
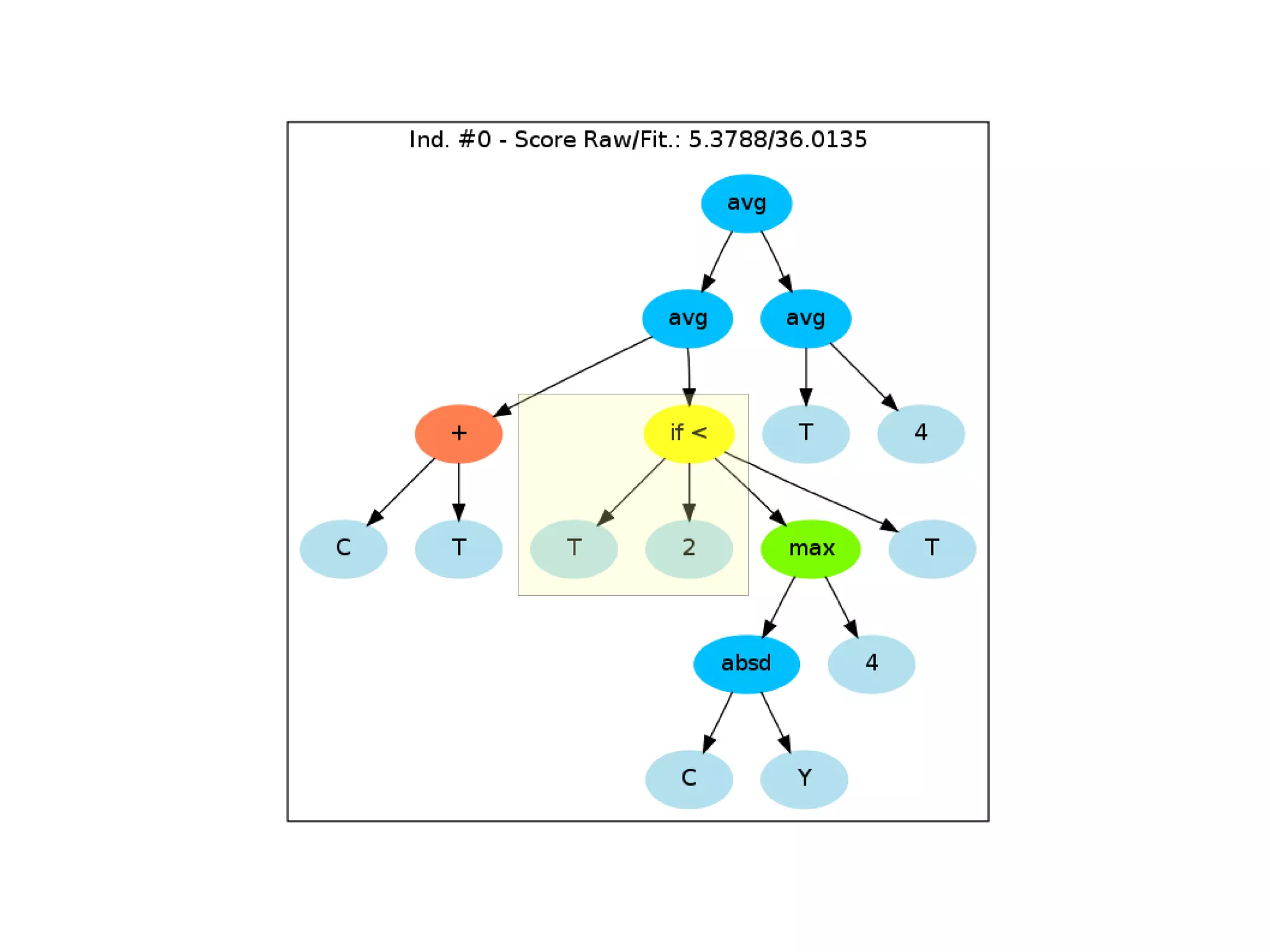
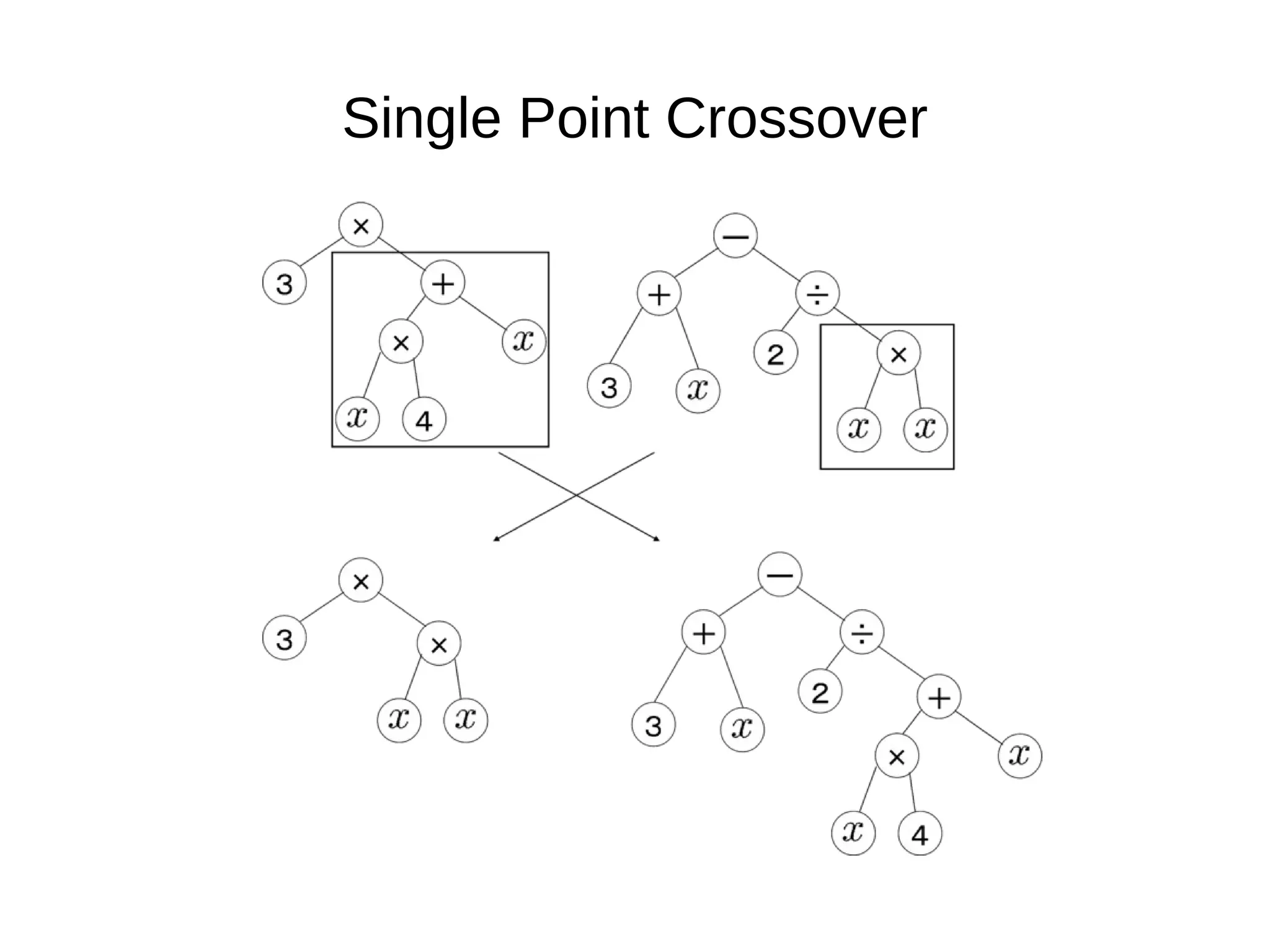
![Initialize The Engine ga = GSimpleGA.GSimpleGA(genome) ga.setParams(gp_terminals = ['per', 'perlast', 'clm'], gp_function_prefix = "gp") ga.setMinimax(Consts.minimaxType["minimize"]) ga.setGenerations(50) ga.setCrossoverRate(1.0) ga.setMutationRate(0.25) ga.setPopulationSize(800)](https://image.slidesharecdn.com/presentation-100811152315-phpapp02/75/Genetic-Programming-in-Python-81-2048.jpg)
![Initialize The Engine ga = GSimpleGA.GSimpleGA(genome) ga.setParams(gp_terminals = ['per', 'perlast', 'clm'], gp_function_prefix = "gp") ga.setMinimax(Consts.minimaxType["minimize"]) ga.setGenerations(50) ga.setCrossoverRate(1.0) ga.setMutationRate(0.25) ga.setPopulationSize(800)](https://image.slidesharecdn.com/presentation-100811152315-phpapp02/75/Genetic-Programming-in-Python-82-2048.jpg)
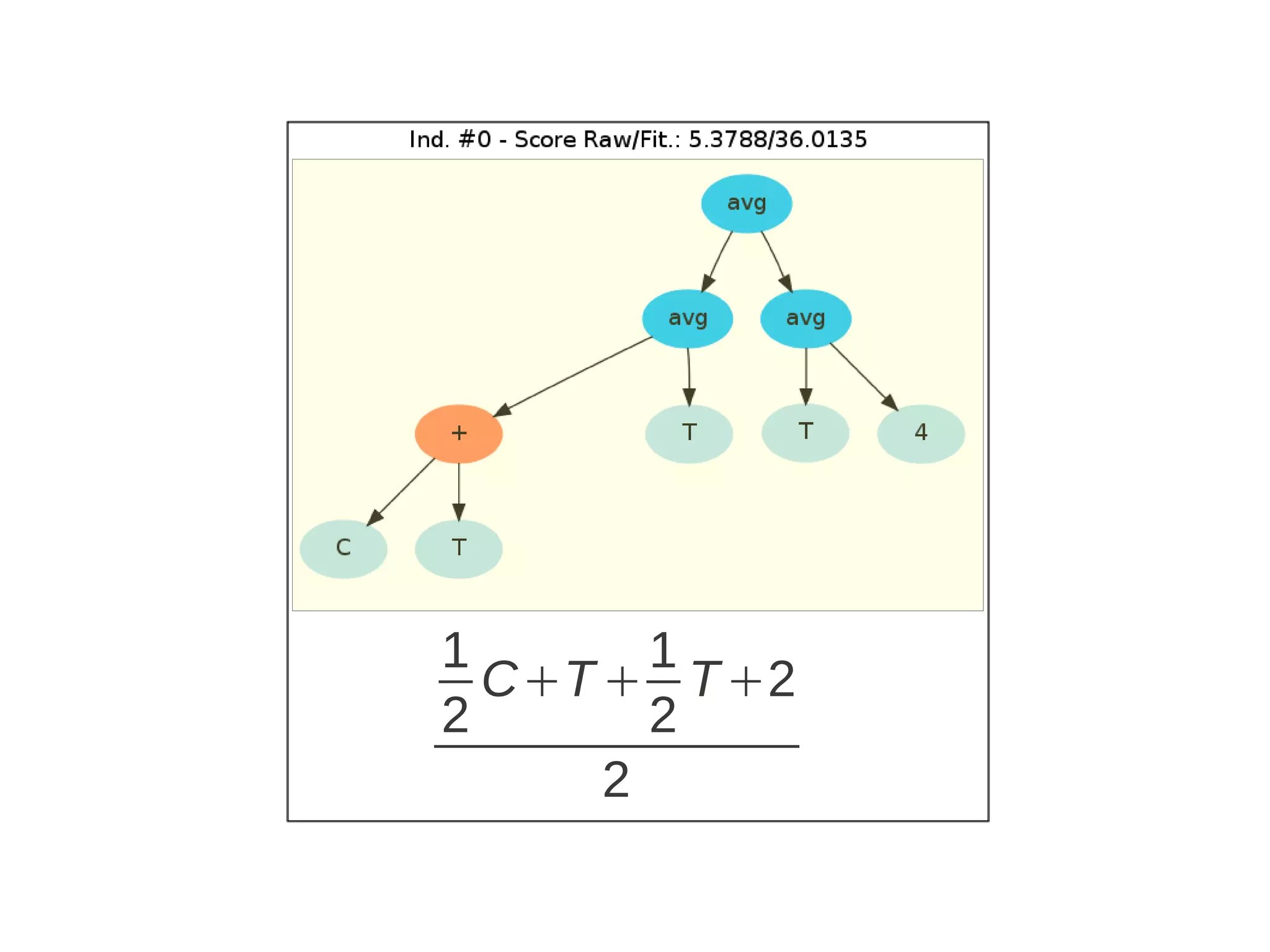
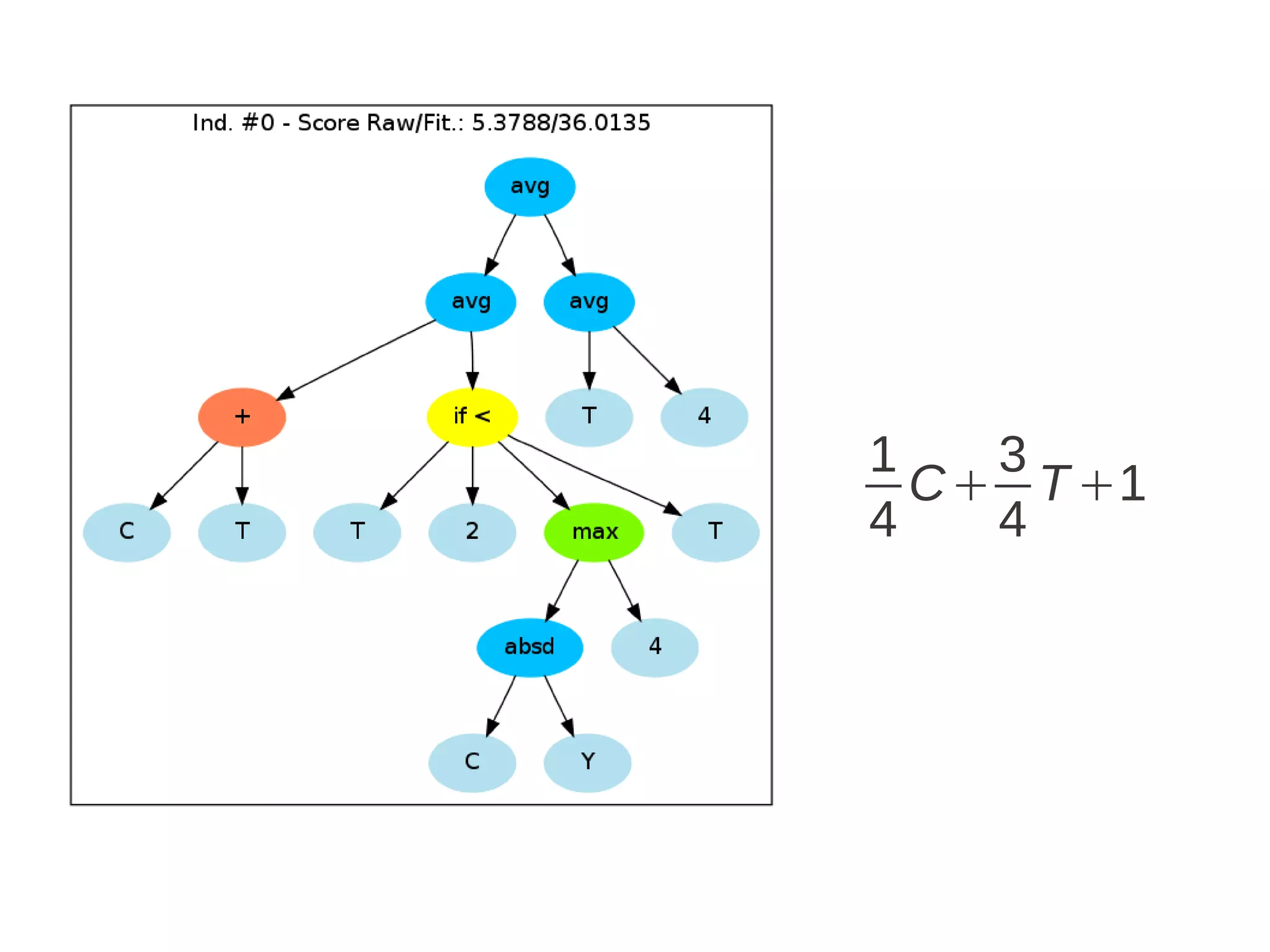


![Run ga = GSimpleGA.GSimpleGA(genome) ga.setParams(gp_terminals = ['per','perlast','clm'], gp_function_prefix = "gp") ga.setMinimax(Consts.minimaxType["minimize"]) ga.setGenerations(50) ga.setCrossoverRate(1.0) ga.setMutationRate(0.25) ga.setPopulationSize(800) ga.evolve(freq_stats=10) best = ga.bestIndividual() print best](https://image.slidesharecdn.com/presentation-100811152315-phpapp02/75/Genetic-Programming-in-Python-87-2048.jpg)