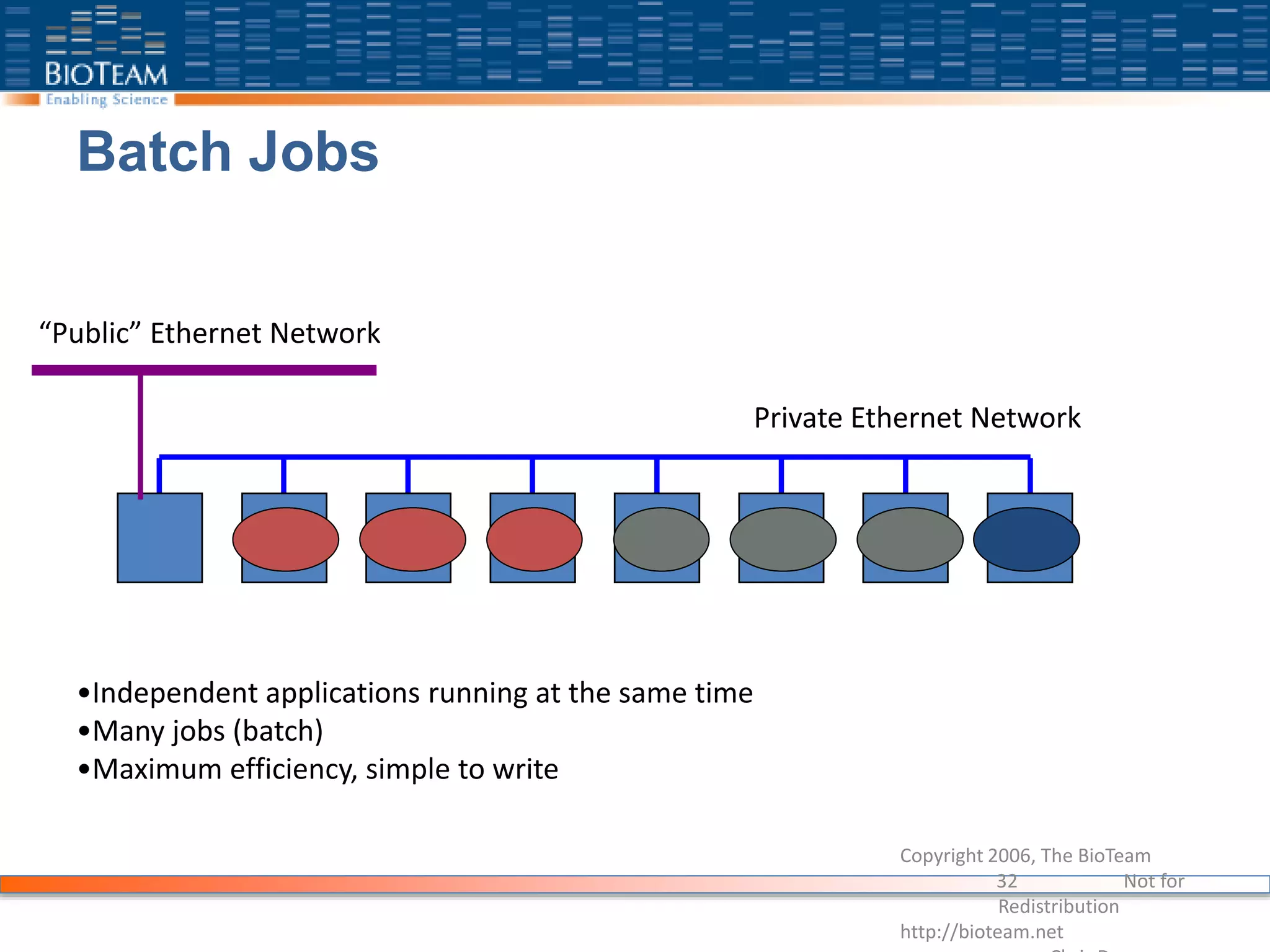
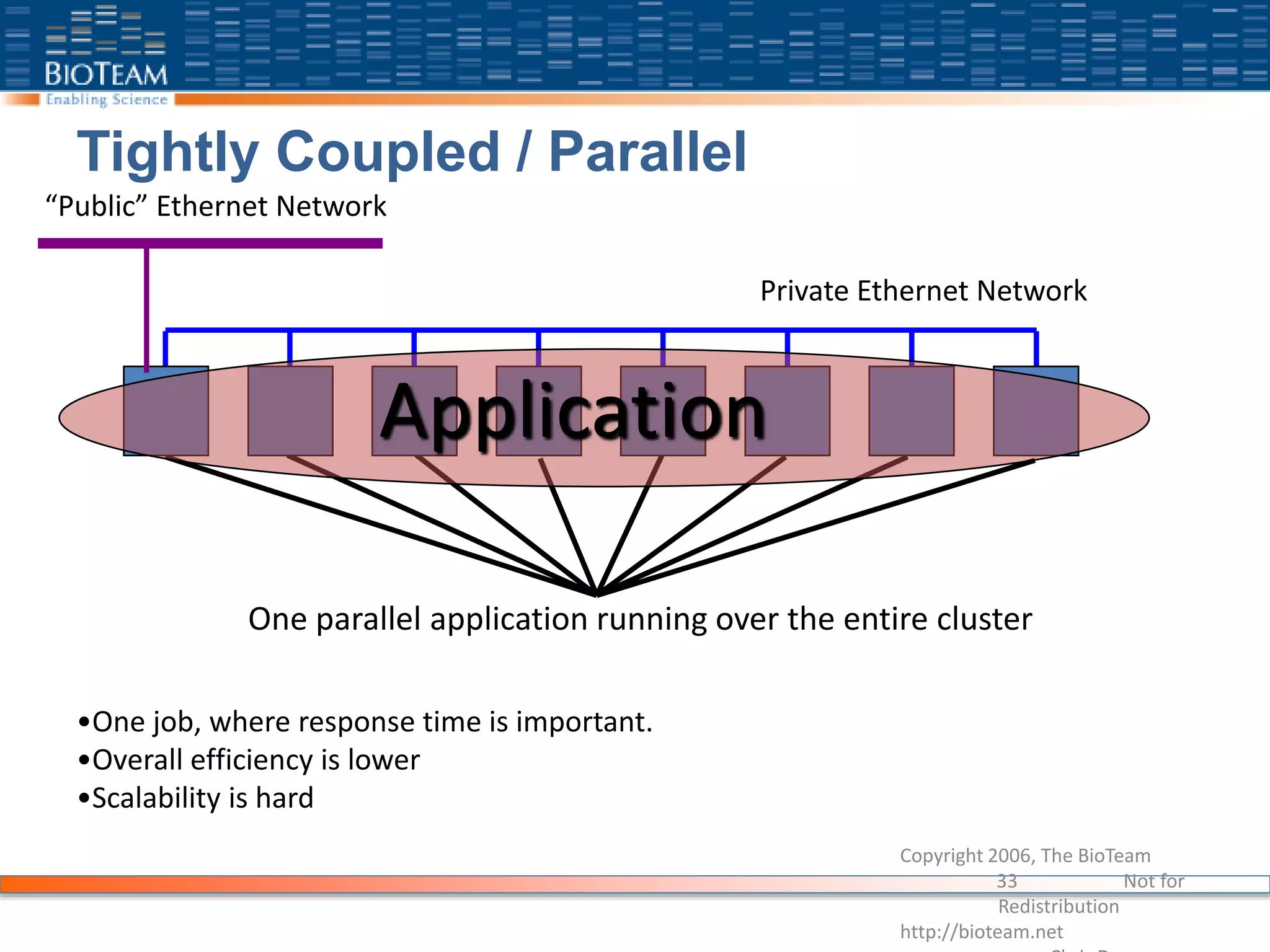
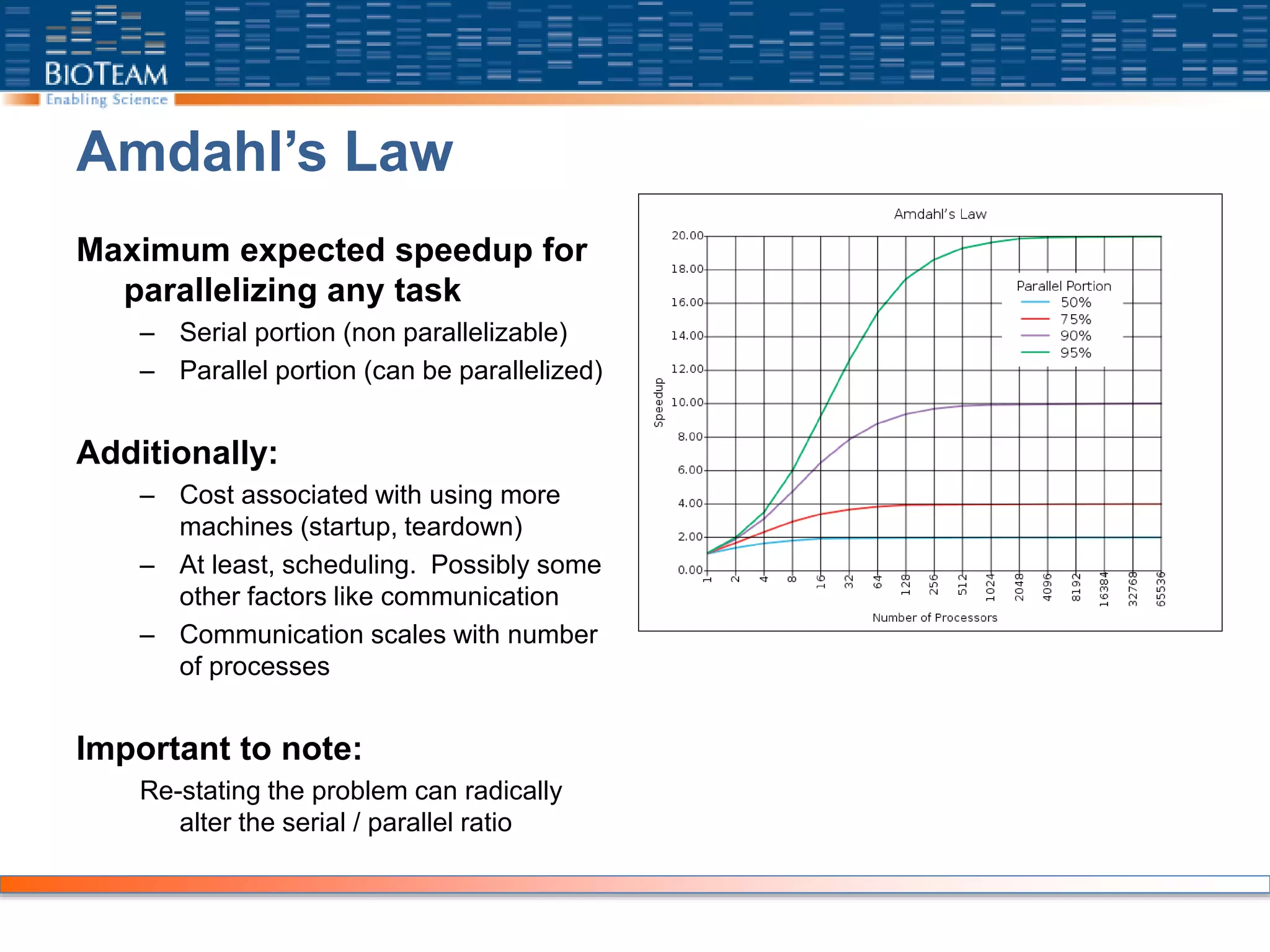
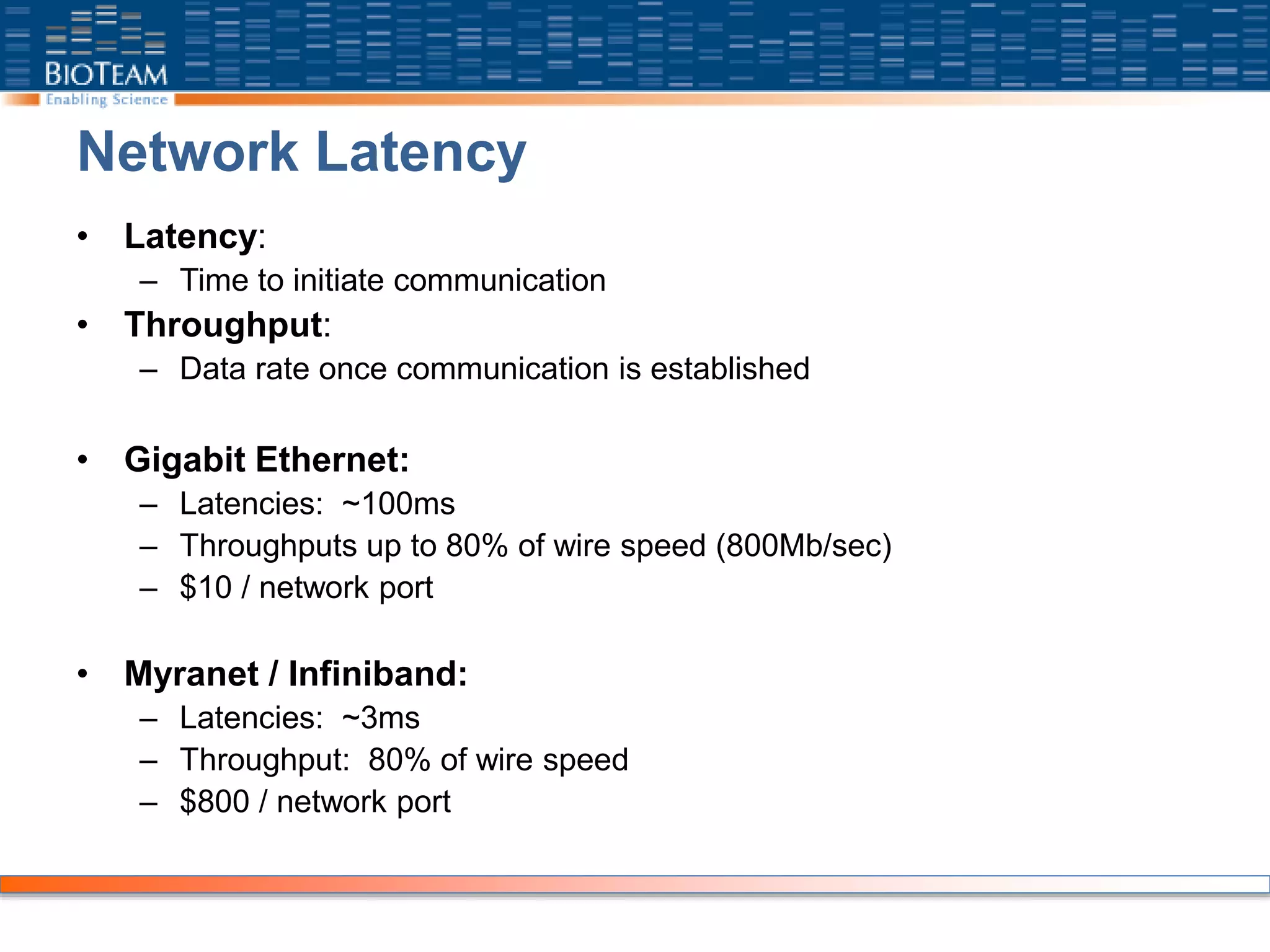
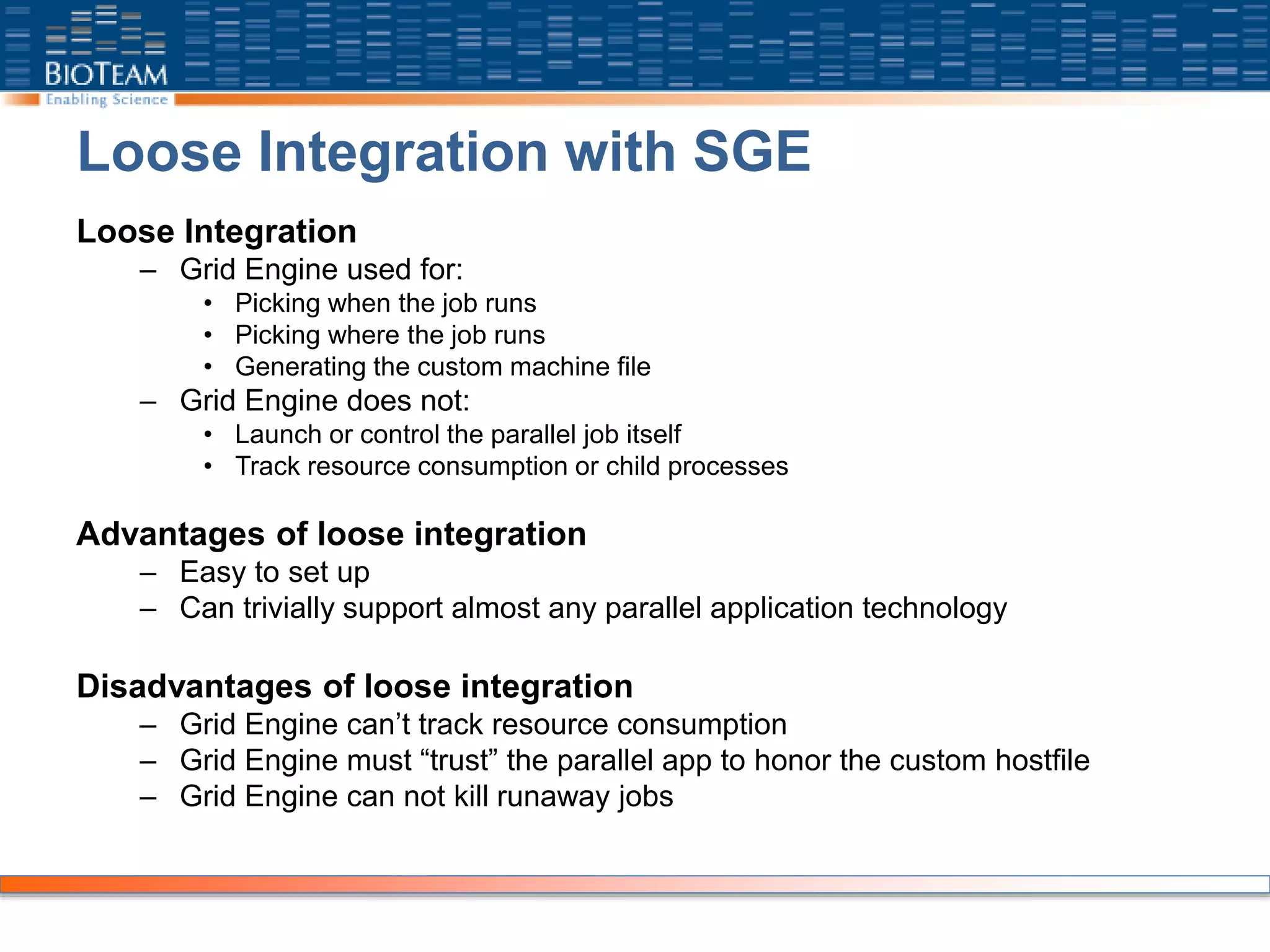
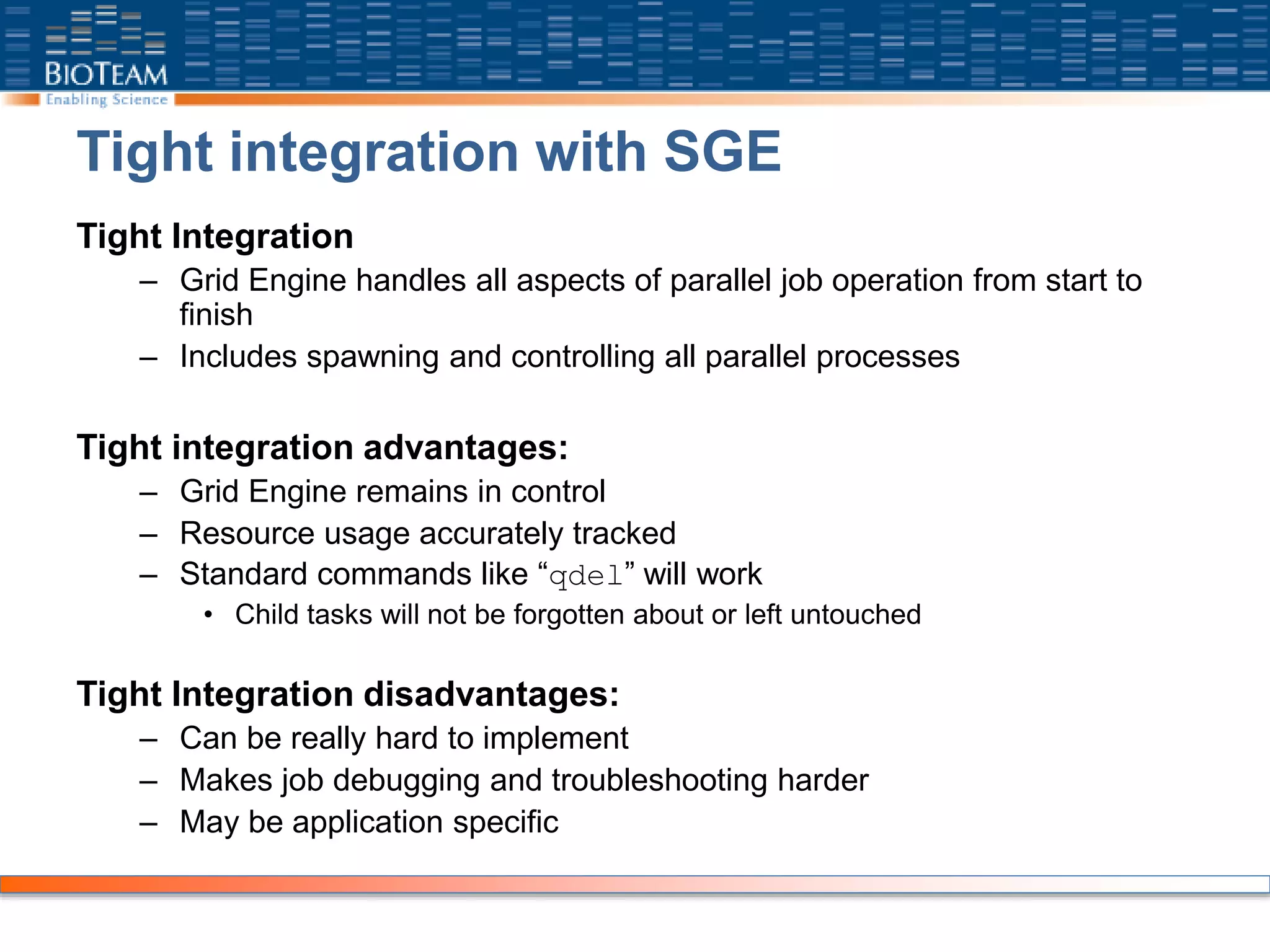
The document provides a comprehensive guide for training users on transitioning from basic Unix commands to running parallel jobs under the Sun Grid Engine (SGE). It includes essential Unix commands, scripting with Bash, submitting and managing jobs in SGE, and concepts of parallel computing. It emphasizes practical exercises, command usage, and performance considerations when working with parallel jobs in a high-performance computing environment.










![Interactive Sessions
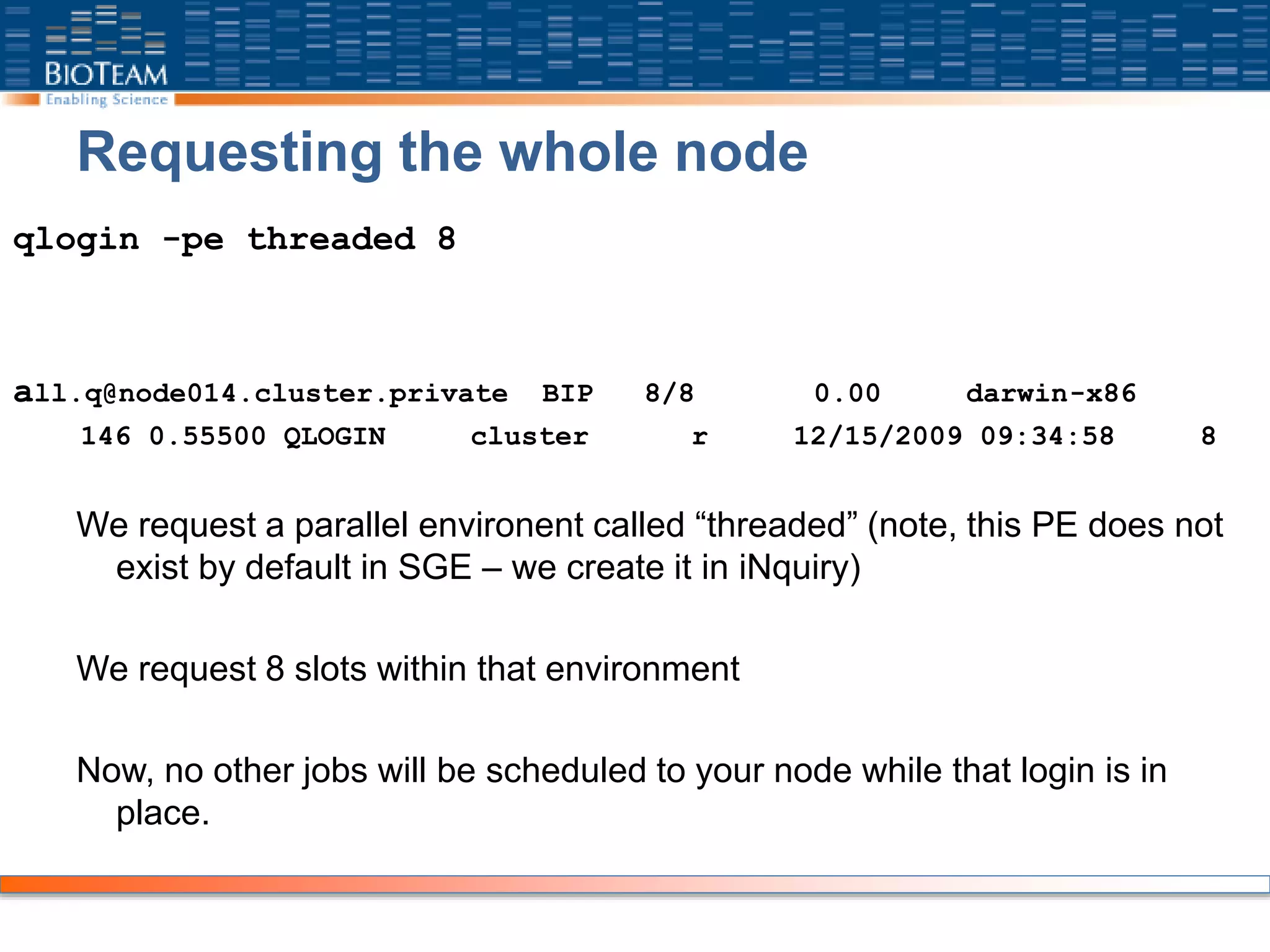
To generate an interactive session, scheduled on any node:
“qlogin”
applecluster:~ cluster$ qlogin
Your job 145 ("QLOGIN") has been submitted
waiting for interactive job to be scheduled ...
Your interactive job 145 has been successfully scheduled.
Establishing /common/node/ssh_wrapper session to host node002.cluster.private ...
The authenticity of host '[node002.cluster.private]:50726 ([192.168.2.2]:50726)'
can't be established.
RSA key fingerprint is a7:02:43:23:b6:ee:07:a8:0f:2b:6c:25:8a:3c:93:2b.
Are you sure you want to continue connecting (yes/no)? yes
Warning: Permanently added '[node002.cluster.private]:50726,[192.168.2.2]:50726'
(RSA) to the list of known hosts.
Last login: Thu Dec 3 09:55:42 2009 from portal2net.cluster.private
node002:~ cluster$
all.q@node002.cluster.private BIP 1/8 0.00 darwin-x86
145 0.55500 QLOGIN cluster r 12/15/2009 09:15:08](https://image.slidesharecdn.com/2009clusterusertraining-200430000751/75/2009-cluster-user-training-14-2048.jpg)


















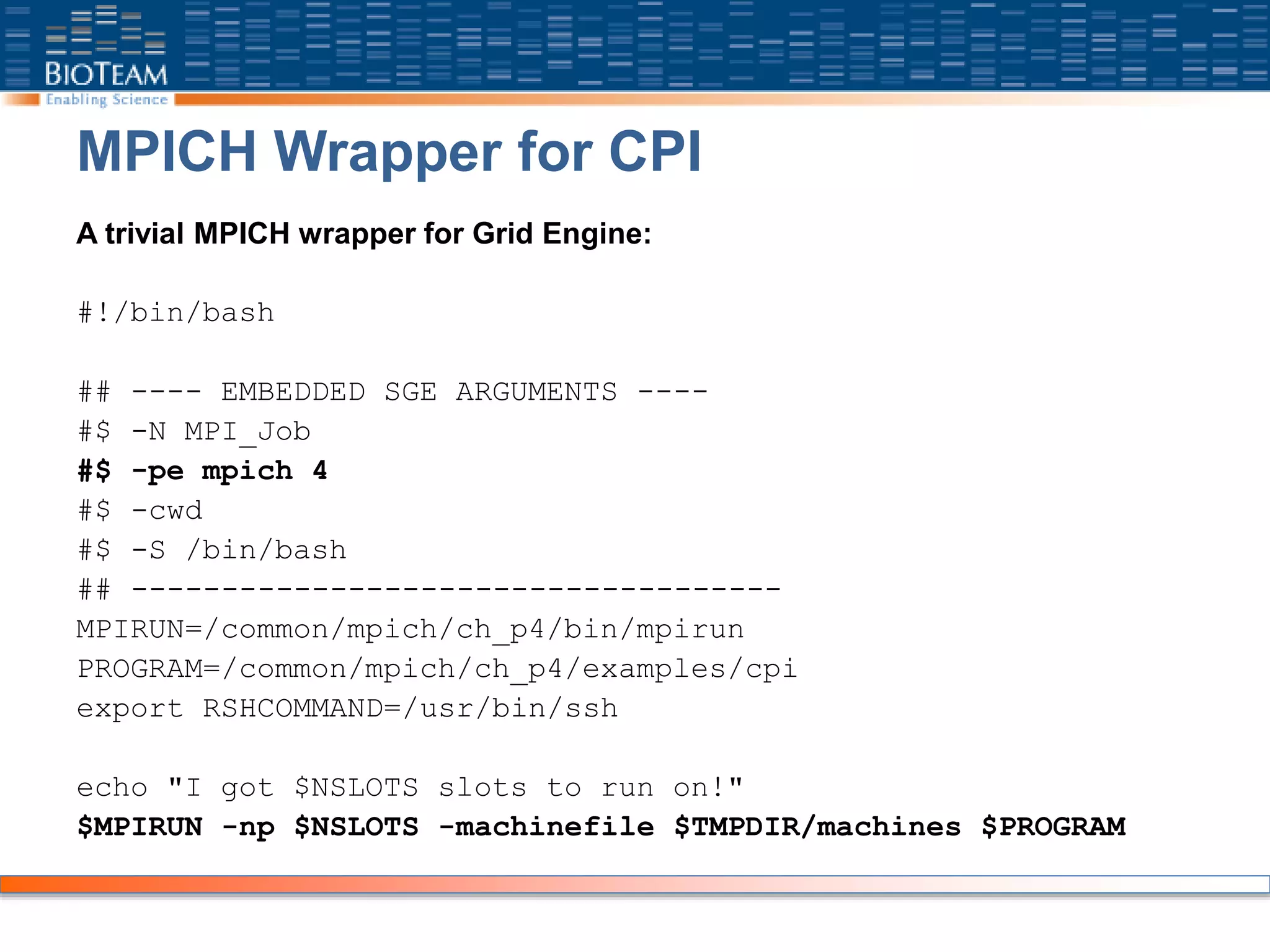
![Job Execution
Submit just like any other SGE job:
[genesis2:~] bioteam% qsub submit_cpi
Your job 234 ("MPI_Job") has been submitted
Output files generated:
[genesis2:~] bioteam% ls -l *234
-rw-r--r-- 1 bioteam admin 185 Dec 15 20:55 MPI_Job.e234
-rw-r--r-- 1 bioteam admin 120 Dec 15 20:55 MPI_Job.o234
-rw-r--r-- 1 bioteam admin 52 Dec 15 20:55 MPI_Job.pe234
-rw-r--r-- 1 bioteam admin 104 Dec 15 20:55 MPI_Job.po234](https://image.slidesharecdn.com/2009clusterusertraining-200430000751/75/2009-cluster-user-training-45-2048.jpg)
![Output
[genesis2:~] bioteam% more MPI_Job.o234
I got 5 slots to run on!
pi is approximately 3.1416009869231245, Error is
0.0000083333333314
wall clock time = 0.001697
[genesis2:~] bioteam% more MPI_Job.e234
Process 0 on node006.cluster.private
Process 1 on node006.cluster.private
Process 4 on node004.cluster.private
Process 2 on node013.cluster.private
Process 3 on node013.cluster.private](https://image.slidesharecdn.com/2009clusterusertraining-200430000751/75/2009-cluster-user-training-46-2048.jpg)