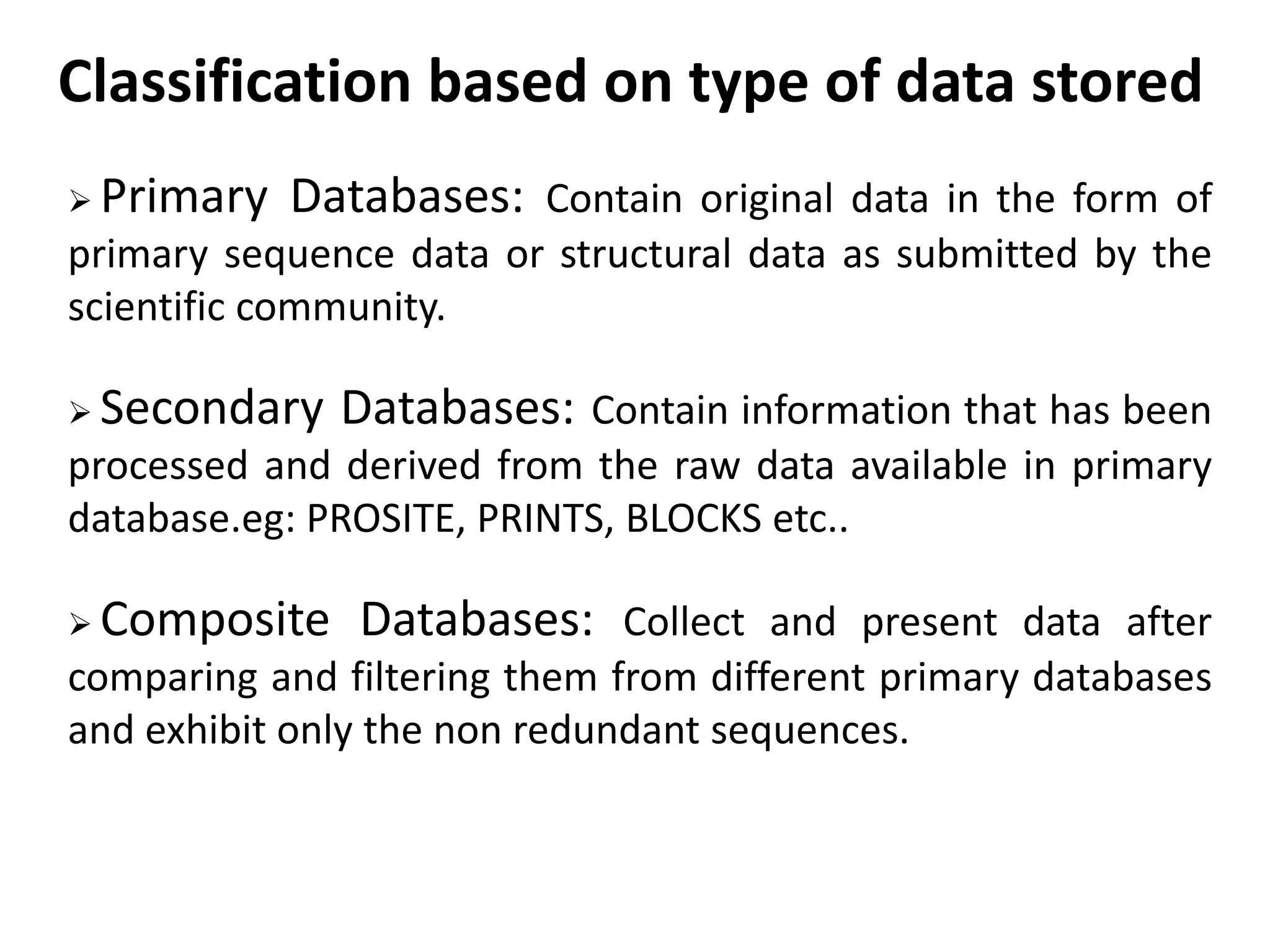
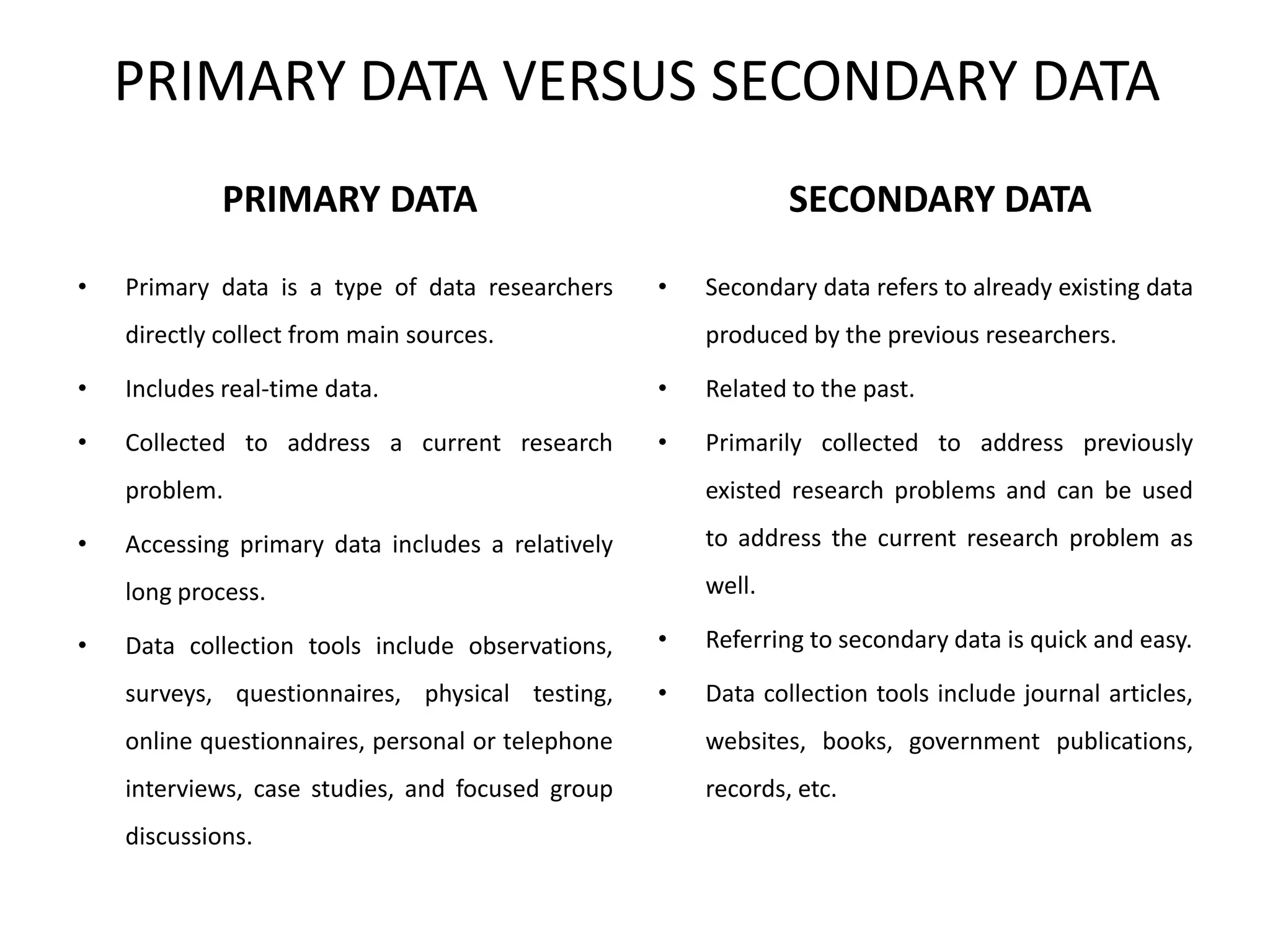
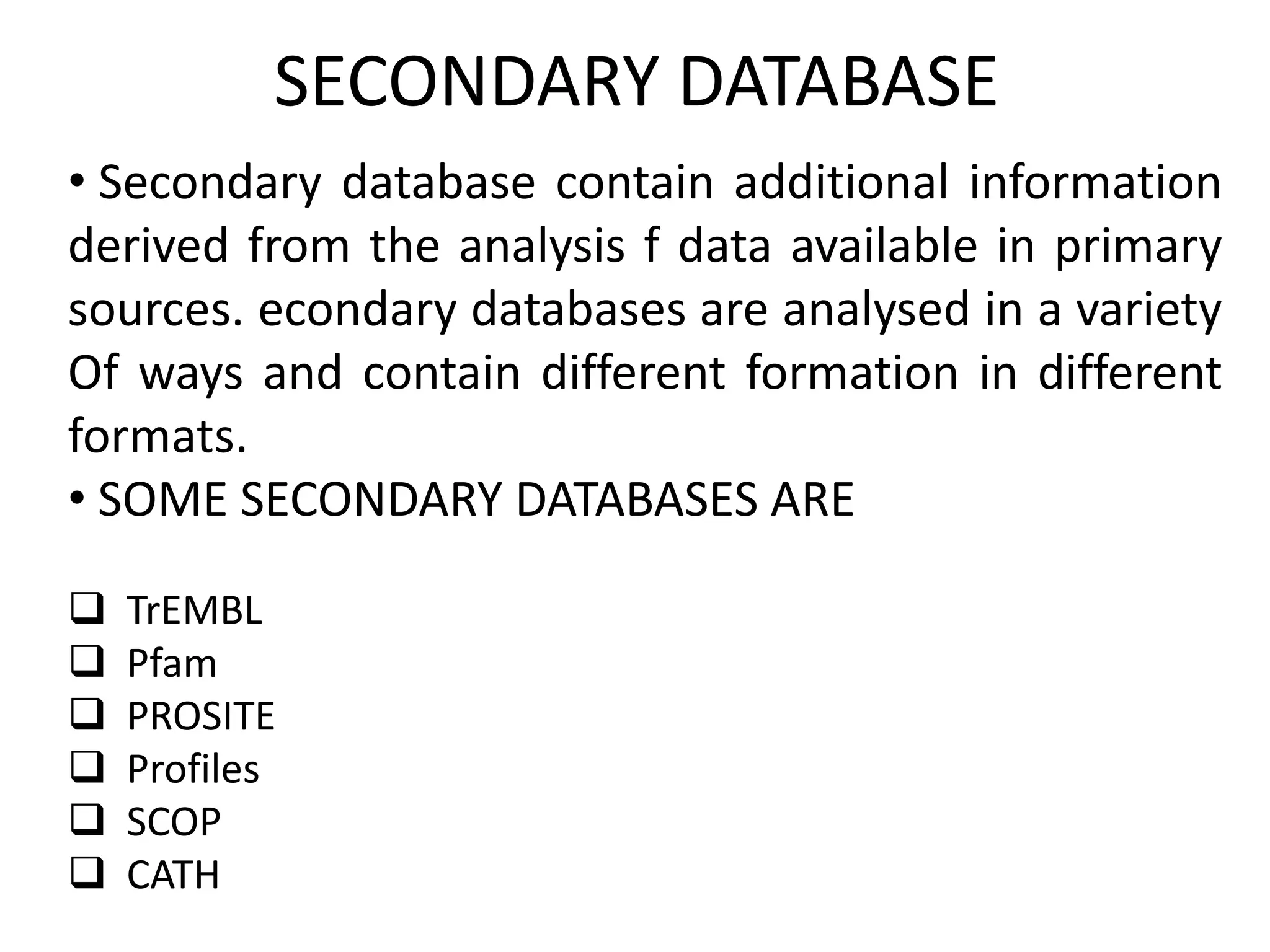
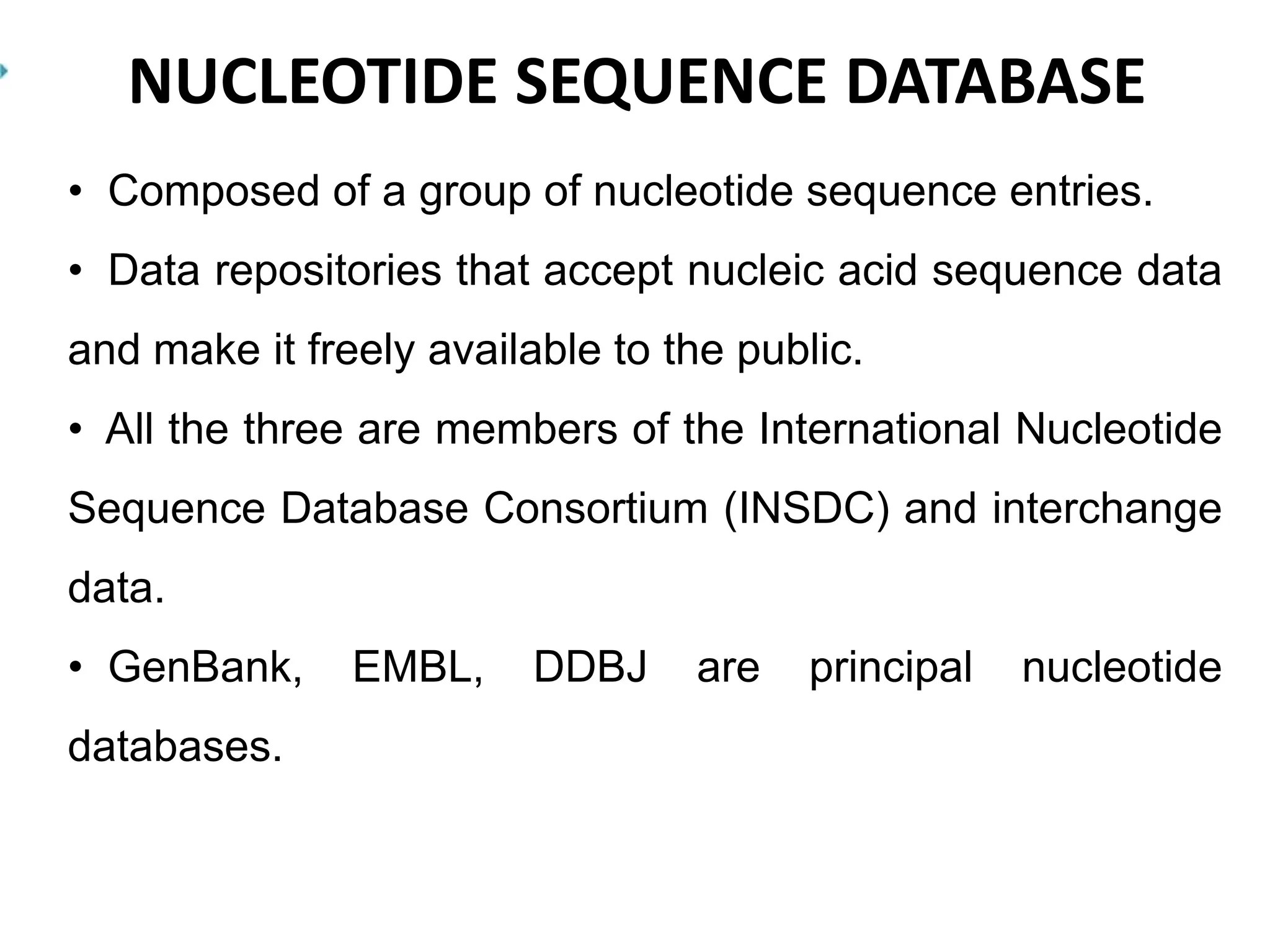
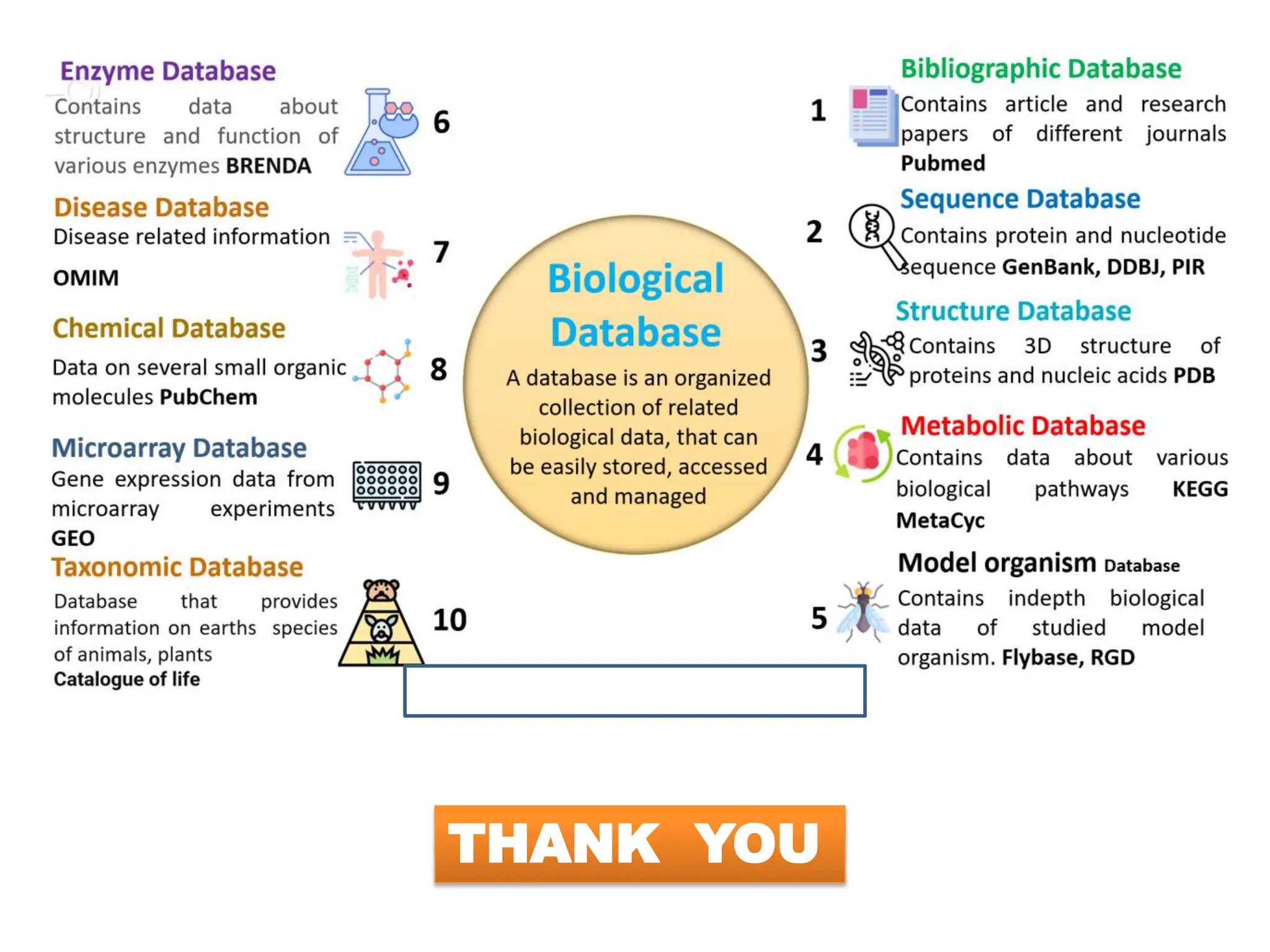
The document discusses biological databases in bioinformatics, highlighting their role in storing, managing, and distributing biological data in a structured and searchable manner. It distinguishes between primary databases, which contain original data, and secondary databases, which provide processed information derived from primary data. Key examples of these databases include GenBank, EMBL, and DDBJ for nucleic acids, and Swiss-Prot and UniProt for protein sequences.